Last Modified
1 January 2025
![]()
分子機能研究所 | Institute of Molecular Function
Latest News Archives
2024/10/12 The research results of the MFDD in silico drug design and discovery research carried out by the Institute for Molecular Function Research, ``Searching for cancer therapeutic drug candidate compounds targeting olfactory receptors using docking screening,'' were presented from he University of Tokyo at the Kanto region meeting of the Japanese Pharmacological Society
2024/09/22 Confirmed the compatibility of HyperChem Release 8 Pro for Windows, as well as Homology Modeling Professional for HyperChem andDocking Study with HyperChem, to the latest version (24H2) of Windows 11 Pro x64.
2024/07/09 A letter of presentation was presented to Motonori Tsuji, representative of the Institute for Molecular Function, at the Yokohama Industrial Association Academic Research Grant Presentation Ceremony.
2024/06/26 Institute of Molecular Function was introduced in "Strengths of in silico drug discovery technology" in the CCS of the Chemical Daily newspaper.
2024/06/11 ``Evaluation of molecular docking calculations using ab initio dynamics software for drug discovery applications'' was selected for the 2024 Yokohama Industrial Association Research and Development Grant, ``Research and Development Grants for Industry-Academia Collaboration''
2024/06/09 Research results including MFDD in silico drug design and discovery contract research conducted by the Institute for Molecular Function Research will be presented by the University of Tokyo at the 45th Annual Meeting of the Japanese Society for Inflammation and Regenerative Medicine (July 17, Fukuoka)
2024/04/04 We are signed an agency agreement with Rikaken Co., Ltd. (Headquarters: Hongo, Bunkyo-ku, Tokyo) for MFDD in silico drug discovery contract research services.
2024/04/01 Motonori Tsuji (Principal Investigator), Yokohama National University, Ochanomizu University, and Tokyo Medical and Dental University have been adopted to the 2024th Research Center for Biomedical Engineering (RCBI) project. Structure-based Drug Design for Retinoic Acid Receptor gamma subtype.
2023/12/06 MFDD In Silico Drug Design Research Service: Our research article (Hydroxyhexylitaconic acids as potent IMP-type metallo-beta-lactamase inhibitors for controlling carbapenem resistance in Enterobacterales) with Nagoya University and Shubun University has been accepted for publication in Microbiology Spectrum (impact factor: 9.043).
2023/11/17 Confirmed the compatibility of HyperChem Release 8 Pro for Windows, as well as Homology Modeling Professional for HyperChem andDocking Study with HyperChem, to the latest version (23H2) of Windows 11 Pro x64.
2023/11/07 Our news regarding to in silico drug design has been published in CCSnews.
2023/10/23 Press Release: Institute of Molecular Function discovered drug candidate using in silico screenings.
2023/08/10 Motonori Tsuji speaks about the Concept and Usages of AutoDock and AtoDock Vina: Structure-based Drug Design using only open free software at Hokkaido University of Scinece.
2023/07/01 Institute of Molecular Function: 20th Anniversary.
2023/06/29 Our news has been published in the Chemical Daily newspaper.
2023/06/16 MFDD In Silico Drug Design Research Service: Our research article (Functional evaluation of rare OASL variants by analysis of SLE patient-derived iPSCs) with Tokyo University has been accepted for publication in Journal of Autoimmunity (impact factor: 14.511).
2023/06/05 Press Release: MFDD In Silico Drug Design Research Service
2023/06/01 - 2023/09/30 20th Anniversary MFDD In Silico Drug Design Research Service
2023/05/26 Our research will be spoken in the 66th Spring Meeting of the Japanese Society of Periodontology.
2023/04/01 Motonori Tsuji (Principal Investigator), Yokohama National University, Ochanomizu University, and Tokyo Medical and Dental University have been adopted to the 2023th Research Center for Biomedical Engineering (RCBI) project. Structure-based Drug Design for Retinoic Acid Receptor gamma subtype.
2023/03/01 - 2023/05/31 20th Anniversary MFDD In Silico Drug Design Research Service
2023/02/10 Minor update of DSHC and HMHC
2022/11/27 Our researches for COVID-19 inhibitions will be reported at the 1st Japan Saliva Care Association.
2022/11/10 Motonori Tsuji will speak about the Development of the COVID-19 Drug Candidates Using In Silico Screening and CADD at the 50th Structure Activity Relationship Symposium.
2022/10/22 Confirmed the compatibility of HyperChem Release 8 Pro for Windows, as well as Homology Modeling Professional for HyperChem andDocking Study with HyperChem, to the latest version (22H2) of Windows 11 Pro x64.
2022/09/27 Our news regarding to COVID-19 has been published in CCSnews.
2022/09/21 Press Release: the Structure-based drug design for the COVID-19 drug candidates.
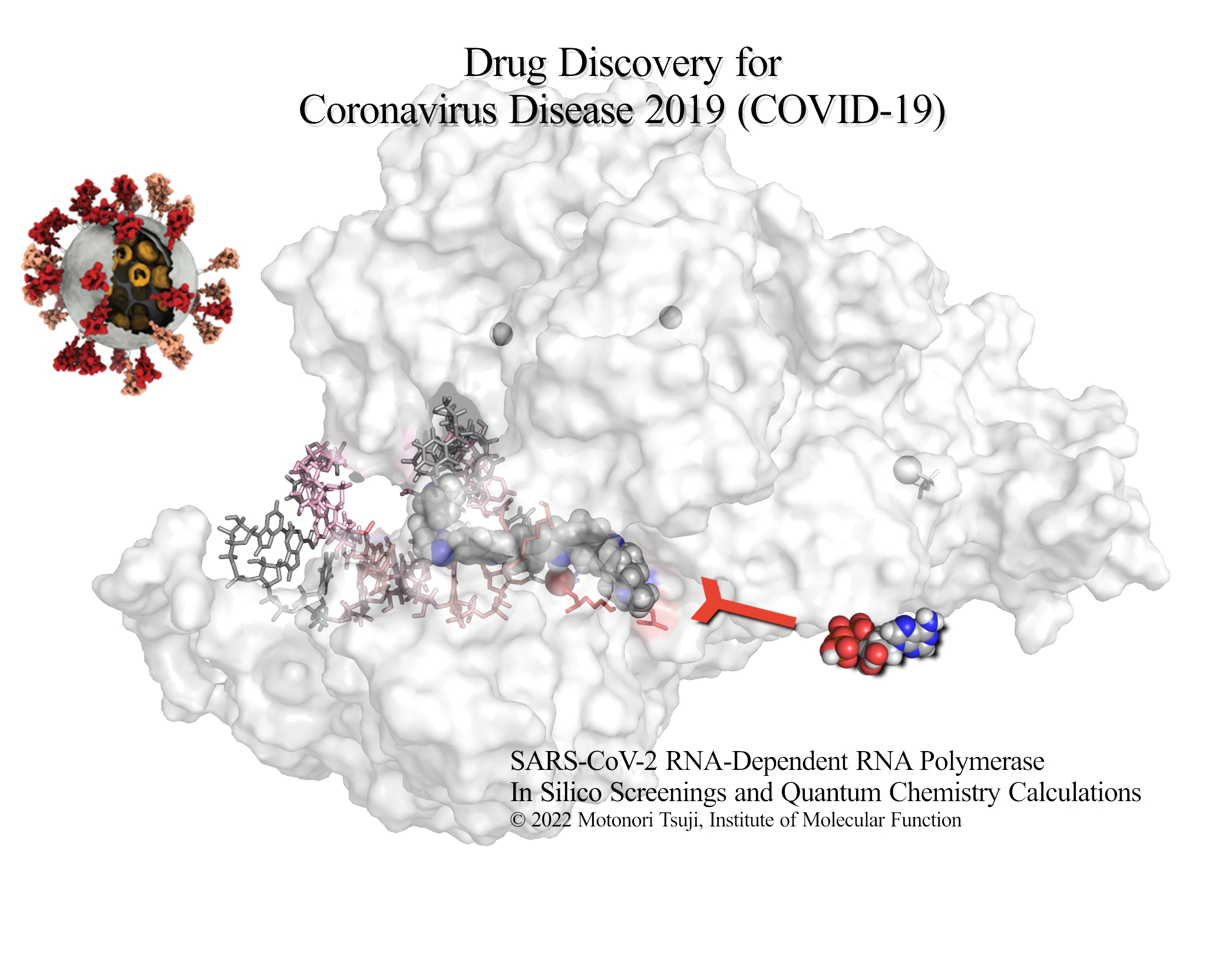
2022/09/20 Motonori Tsuji's article for the COVID-19 drug candidates has been published. Virtual screening and quantum chemistry analysis for SARS-CoV-2 RNA-dependent RNA polymerase using the ChEMBL database: Reproduction of the remdesivir-RTP and favipiravir-RTP binding modes obtained from cryo-EM experiments with high binding affinity. Int. J. Mol. Sci. 23, 11009, 2022.
2022/09/17 Motonori Tsuji's article for the COVID-19 drug candidates has been accepted for publication at International Journal of Molecular Sciences.

2022/06/29 Our news has been published in the Chemical Daily newspaper.
2022/06/15 Our researches for COVID-19 inhibitions reported at the IADR/APP General Session.
2022/06/16 Article fo Motonori Tsuji for the anti-SARS-CoV-2 drug candidates has been selected to the "Top Downloaded Article" by Wiley editorial office.

2022/06/15 Our researches for COVID-19 inhibitions reported at the EuroPrio10.
2022/05/01 Motonori Tsuji (Principal Investigator ), Yokohama National University, Ochanomizu University, and Tokyo Medical and Dental University have been adopted to the 2022th Research Center for Biomedical Engineering (RCBI) project. Structure-based Drug Design for RARgamma Atagonists.
2022/03/04 Our researches for the Structure-based Drug Design of COVID-19 at the Research Center for Biomedical Engineering.
2022/02/11 Our researches reported at the 5th Annual Scientific Meeting of the Japanese Society for Immunodeficiency and Autoinformmatory Diseases.
2021/11/07 Confirmed the compatibility of HyperChem Release 8 for Windows, Homology Modeling Professional for HyperChem, and Docking Study with HyperChem to the latest version (21H2) of Windows11 Pro.
2021/10/09-2021/10/11 Our researches for COVID-19 inhibitions have been reported at the 63th Japanese Association for Oral Biology.
2021/10/04 Press Release: Inhibition of main infective pathway of SARS-CoV-2 in oral cavity
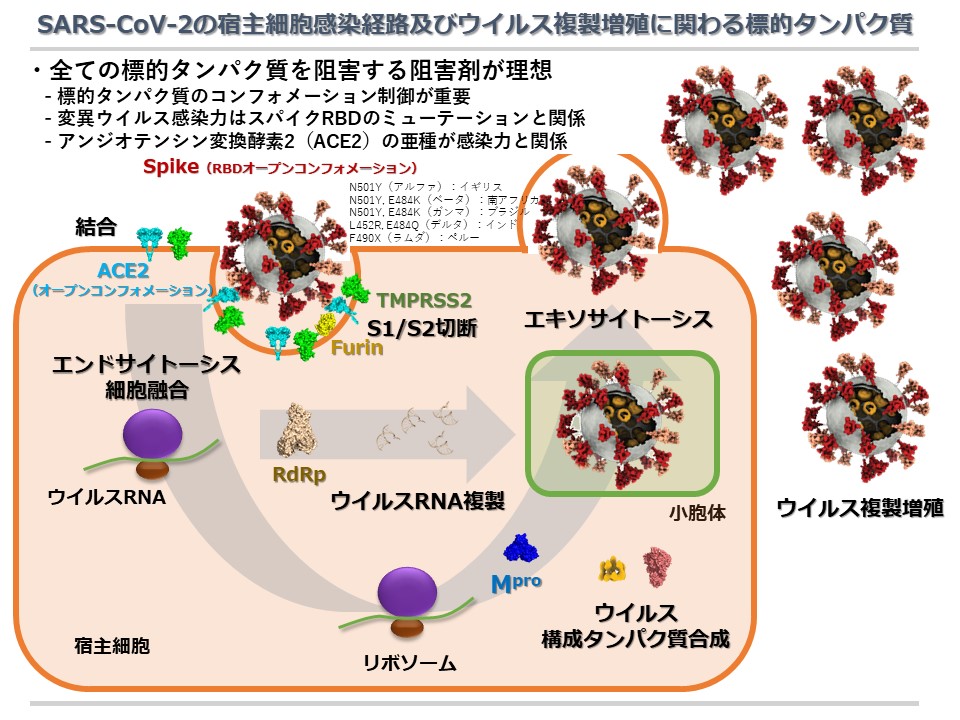
2021/10/01 Our research for COVID-19 inhibitions has been reported at the 14th International Conference of Asian Academy of Preventive Density.
2021/09/03 Our research article for COVID-19 inhibition has been accepted for publication in PLOS ONE.
2021/06/30 Our news has been published in the Chemical Daily newspaper.
2021/06/23 Motonori Tsuji has contracted the corporate research regarding to retinoids with ChemPhys Co. LTD.
2021/05/19 Confirmed the compatibility of HyperChem Release 8 for Windows, Homology Modeling Professional for HyperChem, and Docking Study with HyperChem to the latest version (21H1) of Windows10 Pro x64.
2021/05/14 Motonori Tsuji has been invited to speech at Business Research Institute, 34th CAMM Forum. Development and Application of Structure-based Drug Design Technology - Drug Design for Nuclear Receptor Ligands and COVID-19 medicines.
2021/04/01 Motonori Tsuji (Principal Investigator ), Osaka University, Ochanomizu University, and Tokyo Medical and Dental University have been adopted to the 2021th Research Center for Biomedical Engineering (RCBI) project. Structure-based Drug Design for anti-SARS-CoV-2 drugs.
2021/03/19-2021/03/22 Our researches for COVID-19 inhibitions have been reported at the 101th Chemical Society of Japan.
2021/03/05 Motonori Tsuji will speak about the Structure-based Drug Design for COVID-19 at the Research Center for Biomedical Engineering.
2021/02/18 Research paper of Motonori Tsuji has been nominated for award of FEBS Open Bio journal.
2020/12/22 Motonori Tsuji (Principal Investigator), Osaka University, Ochanomizu University, and Tokyo Medical and Dental University have been adopted to the 2020th Research Center for Biomedical Engineering (RCBI) project. Structure-based Drug Design for anti-SARS-CoV-2 drugs.
2020/12/11 Announced the latest version 8.1.4 of Docking Study with HyperChem and Homology Modeling Professional for HyperChem.
2020/11/11 Confirmed the compatibility of HyperChem Release 8 for Windows, Homology Modeling Professional for HyperChem, and Docking Study with HyperChem to the latest version 20H2 of Windows 10 Pro x64.
2020/10/17 Motonori Tsuji will speak about the Identification of COVID-19 drugs: Virtual Screening for SARS-CoV-2 Main Protease using Nuclear Receptor Ligand Database at the 31th Japanese Society for Retinoid Research.
Motonori Tsuji received the Award of the 31th Japanese Society for Retinoid Research.
2020/09/28-2020/11/30 Our research (together with University of Hyogo and Toyama Prefectural University) for the Coronavirus Protease Inhibition by Phytochemicals and its Metabolites has been reported at Innovation Japan 2020 of Japan Science and Technology Agency.
2020/09/15 Our resaerch article for acetylcholine esterase inhibitor has been accepted for publication in Chem. Pharm. Bull. journal.
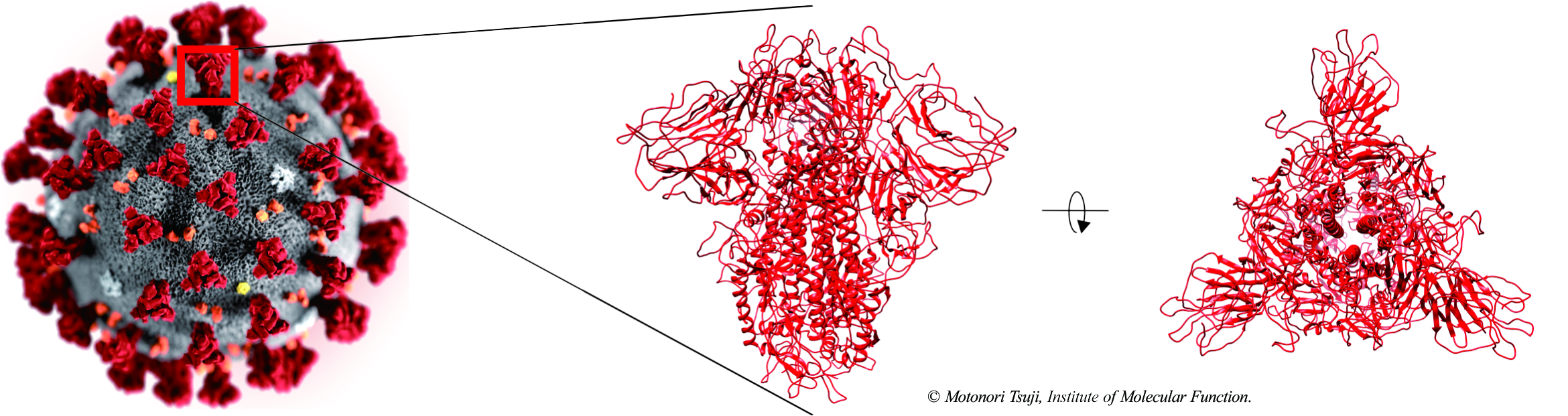
2020/08/14 Protein-protein docking simulations between SARS-CoV-2 spike protein and human ACE2
2020/07/16 Our news has been published in the Chemical Daily newspaper.
2020/07/02 Our research article (together with Osaka University) for chikungunya virus antibody has been accepted for publication in J. Virol.
2020/05/09 Our news regarding to COVID-19 has been published in CCSnews.
2020/05/05 Fragment molecular orbital quantum chemistry calculations of SARS-CoV-2 main protease
2020/05/01 Press Release: Publication of list of potential anti-SARS-CoV-2 drug candidates
Published the article for the potential anti-SARS-CoV-2 drug candidates using in silico screening. Potential anti-SARS-CoV-2 drug candidates identified through virtual screening of the ChEMBL database for compounds that target the main coronavirus protease. FEBS Open Bio, DOI:10.1002/211-5463.12875.
2020/04/27 Nano-second scale molecular dynamics simulations for SARS-CoV-2 main protease apo form
2020/03/07 In silico screenings targeting SARS-CoV-2 (2019-nCoV) main protease (Japanease).

2020/02/25 Antibody design targeting 2019-nCoV spike glycoprotein (Japanease).
2019/12/11 Customer's research article, which Molecular Function has supported the study, has been accepted for publication in J. Pharmacol. Exp. Ther.
2019/11/15 Published the Molecular Function's Loadmap.
2019/11/15 Announced the latest version 8.1.3.
2019/11/13 Confirmed that HyperChem Release 8 for Windows, Homology Modeling Professional for HyperChem, and Docking Study with HyperChem are compatible to the latest Windows version (Windows 10 Pro x64, Version 1909).
2019/10/01 Open the Commissioned Calculation Service Web Site (Japanese).
2019/09/01 Research results of MFDD service will be demonstrated at Asian-African Research Forum on Emerging and Reemerging Infections 2019 (2019/09/05-06).
2019/06/17 Our news has been published in the Chemical Daily newspaper.
2019/06/01 Confirmed that HyperChem Release 8 for Windows, Homology Modeling Professional for HyperChem, and Docking Study with HyperChem are compatible to the latest Windows version (Windows 10 Pro x64 2019, Version 1903).
2019/02/08 Revised the home pages for mobile devices.
2019/01/26 Open the MFDD Service Web Site (Japanese).
2018/07/01 15th Anniversary
2018/06/07-08 Motonori Tsuji will speak a research entitled "Advancement of the Current Drug Design Technologies via In Silico Drug Design System, Docking Study with HyperChem (DSHC) and Homology Modeling Professional for HyperChem (HMHC)." at the 2018 Spring annual meeting of Society of Computer Chemistry, Japan.

2018/03/27 Motonori Tsuji will speak a research entitled "Antagonist-Perturbation Mechanism for Activation Function-2 Fixed Motifs: Active Conformation and Docking Mode of Retinoid X Receptor Antagonists" at the 138th annual meeting of the pharmaceutical society of Japan.

2018/03/21 Sophisticated Molecular Modeling Software, HyperChem site is started in Japan.
2018/03/09 Our news has been published in CCSnews.
2018/02/23 Announcing Docking Study with HyperChem, Revision H1 and Homology Modeling Professional for HyperChem, Revision H1.
2018/01/09 The Latest In Silico Drug Design Platform: Evolution of Docking Study with HyperChem, Revision H1 and Homology Modeling Professional for HyperChem, Revision H1. Coming Soon. (Strengthened quantum mechanics calculation program, Gaussian, ONIOM Interface, compatible to file format of molecular dynamics calculation program, NAMD (VMD: CHARMM-based PDB), compatible to file format of fragment molecular orbital calculation programs, ABINIT-MP (BioStation Viewer) and GAMESS (Fu /Facio), and supported AutoDock Vina Virtual Screening Interface (preparation of compound database - preparation of all input files - execution - viewing and filtering of hits; supports also simultaneous analysis of both Docking Study and AutoDock Vina results))
2017/11/19 Motonori Tsuji spoke a research entitled "Identifying the Receptor Subtype Selectivity of Retinoids via Docking, QM/MM, and Quantum Mechanics Calculations: ATRA Can Act as an Endogenous RXR Ligand" at the 28th symposium of retinoids of Japan society for retinoid research.
2017/06/21 Our news has been published in the Chemical Daily newspaper.
2017/05/17 Our news has been published in CCSnews.
2017/05/16 Origin of the agonism and antagonism mechanisms in the nuclear receptors via quantum mechanics (using core technologies of Docking Study with HyperChem and Homology Modeling Professional for HyperChem) has been accepted for publication. Motonori Tsuji. J. Comput. Aided Mol. Des. 31, 577-585, 2017, DOI: 10.1007/s10822-017-0025-6.
2017/04/10 Confirmed that the products are compatible to Gaussian16W 64-bit multi-processor version (Revision A.03). I really appreciate great help of Gaussian, Inc.
2017/03/28 All series of HyperChem of Hypercube, Inc. are available from our web site.
2017/03/27 Motonori Tsuji spoke a research entitled "Helix H3 Three-Point Initial-Binding Hypothesis: Understanding of Both the Ligand-Trapping Mechanism and the Simultaneous Structural Transition from Apo-Form to Holo-Form of the Ligand-Binding Domain in the Nuclear Receptor Superfamily" at the 137th annual meeting of the pharmaceutical society of Japan.

2017/03/15 Our news has been published in CCSnews.
2017/02/15 Press Release: Origin of the receptor subtype selectivity via quantum mechanics has been published. Motonori Tsuji, et. al. FEBS Open Bio. 7, 391-396, 2017, DOI: 10.1002/2211-5463.12188.
2016/12/29 Origin of the receptor subtype selectivity via quantum mechanics has been accepted for publication.
2016/06/24 Our news has been published in the Chemical Daily newspaper.
2016/04/16 Confirmed that ONIOM Interface for Receptor and Gaussian Interface for HyperChem were compatible with the latest Gaussian09W (64bit-multiprocessor version, Revision E.01) under support of Gaussian, Inc. I really appreciate great help of Gaussian, Inc.
2016/03/27 Motonori Tsuji spoke a research entitled "Geometrical Dependence of the Highest Occupied Molecular Orbital in Bicyclic Systems: Origin of the Facial Stereoselectivity of Bicyclic Olefins" at the 136th annual meeting of the pharmaceutical society of Japan.
2016/03/26 Announcing Docking Study with HyperChem 2016 and Homology Modeling Professional for HyperChem 2016, Revision G1.
2016/03/26 The function of ONIOM Interface for Receptor was strengthened (compatible to whole system of protein and nucleic acid molecular system).
2015/12/29 Our news has been published in CCSnews.
2015/12/01 Confirmed that Docking Study with HyperChem and Homology Modeling Professional for HyperChem were compatible with Windows 10 Professional (64 bit Version).
2015/11/04 Press Release: Origin of the ligand recognition mechanism in the nuclear receptor superfamily has been published. Motonori Tsuji. J. Mol. Graph. Model. 62, 262-275, 2015.
2015/10/07 The first success of the structural transition of the holo-form formation simulations and the discovery of the driving forces involved in the ligand entry in the nuclear receptor superfamily using Docking Study with HyperChem and Homology Modeling Professional for HyperChem has been accepted for publication.
2015/09/13 A proposed molecular mechanism of ATRA via retinoid X receptors (using core technologies of Docking Study with HyperChem and Homology Modeling Professional for HyperChem) has been accepted for publication. Motonori Tsuji, et. al. J. Comput. Aided Mol. Des. 29, 975-988, 2015.
2015/06/25 Our news has been published in the Chemical Daily newspaper.
2015/05/08 Our news has been published in CCSnews.
2015/04/17 Press Release: Origin of the facial stereoselectivity, which is the most important and unsolved issue in organic chemistry, has recently been accepted for publication. Motonori Tsuji. Asian J. Org. Chem. 4, 659-673, 2015.
2015/03/26 Motonori Tsuji spoke a research entitled "Structure and function of the local motifs in the canonical structure of the ligand-binding domain in the nuclear receptor superfamily" at the 135th annual meeting of the pharmaceutical society of Japan.
2015/01/04 Announcing Docking Study with HyperChem 2015 and Homology Modeling Professional for HyperChem 2015, Revision G1.
2014/10/14 Announcing Docking Study with HyperChem and Homology Modeling Professional for HyperChem, Revision F3.
2014/10/02 Confirmed that Docking Study with HyperChem and Homology Modeling Professional for HyperChem were compatible with Windows 10 Technical Preview (64 bit Version).
2014/03/18 A paper regarding the folding mechanism and the agonism and atagonism mechanism of the ligand-binding domain in the nuclear receptor superfamily has recently been published. Motonori Tsuji J. Struct. Biol. 185, 355-365, 2014.
2014/02/08 Our news has been published in CCSnews.
Contributions for the Computer-Aided Drug Design of Coronavirus Disease 2019 (COVID-19)
Protein-protein docking simulations between SARS-CoV-2 spike protein and human ACE2
Fragment molecular orbital quantum chemistry calculations of SARS-CoV-2 main protease
Nano-second scale molecular dynamics simulations for SARS-CoV-2 main protease apo form
In silico screenings targeting SARS-CoV-2 (2019-nCoV) main protease
Antibody design targeting 2019-nCoV spike glycoprotein
Released the latest version 8.1.5 at Aug 10, 2023
HyperChem can be drastically upgraded as the latest drug discovery system by the Homology Modeling Professional for HyperChem (HMHC) and Docking Study with HyperChem (DSHC) applications.
Announce
What's New in Revision H1. (The latest version is 8.1.5 at Aug 10, 2023)
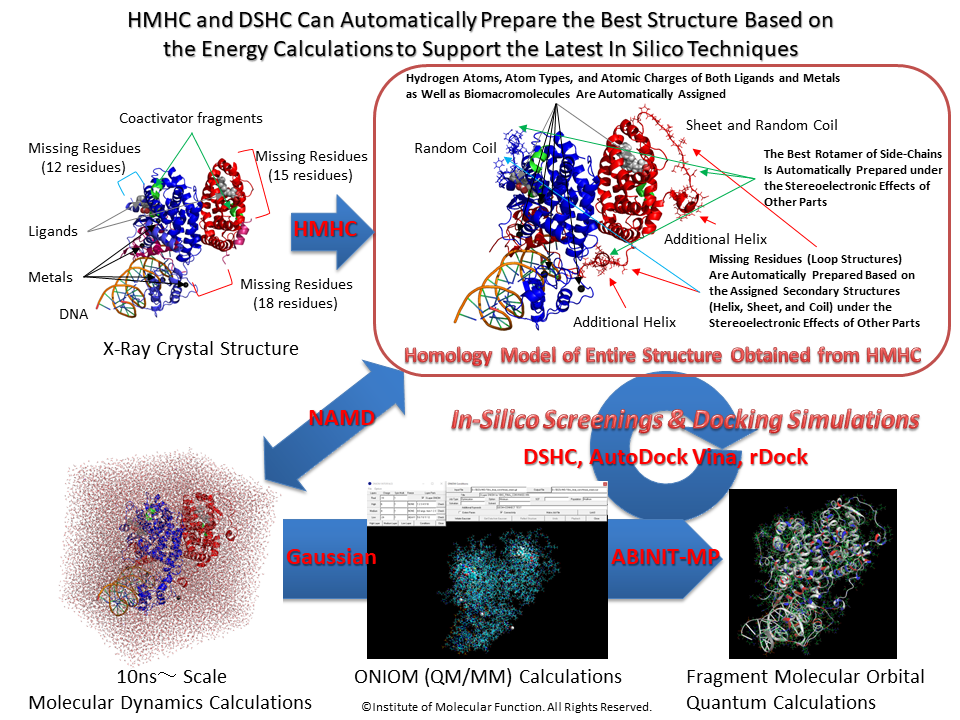
The Latest In Silico Drug Design Platform: Evolution of Docking Study with HyperChem, Revision H1 and Homology Modeling Professional for HyperChem, Revision H1.
Strengthened quantum mechanics calculation program, Gaussian,ONIOM Interface, compatible to file format of molecular dynamics calculation program, NAMD (VMD: CHARMM-based PDB), compatible to file format of fragment molecular orbital calculation programs, ABINIT-MP (BioStation Viewer) and GAMESS (Fu /Facio), and supported AutoDock Vina Virtual Screening Interface (preparation of compound database - preparation of all input files - execution - viewing and filtering of hits; supports also simultaneous analysis of both Docking Study and AutoDock Vina results) and rDock docking screening interface.
In quantum mechanics calculations for the entire structure of big molecular system such as biomacromolecular system, the single point calculations for the initial structure obtained from classical molecular mechanics calculations and/or classical molecular dynamics calculations cannot converge or will give abnormal energies. For obtaining reliable results from quantum mechanics calculations such as fragment molecular orbital methods of entire molecular system, the initial structure must be prepared using geometrical optimization calculations via ONIOM methods. ONIOM Interface for Receptor can provide the best solution for the quantum mechanics calculations of the entire molecular system of which the precise initial structure is prepared from Docking Study with HyperChem and Homology Modeling Professional for HyperChem.
Motonori Tsuji, et. al. FEBS Open Bio., 7, 391-396, 2017, DOI: 10.1002/2211-5463.12188.
Docking Study with HyperChem, Revision H1
with AutoDock Vina In Silico Screenings Interface
Compatible to rDock Docking Simulations, NAMD Molecular Dynamics and ABINIT-MP / GAMESS Fragment Molecular Orbital Quantum Mechanics Calculations for Entire Molecular System
- Carrying The Next-generation Drug Discovery Technology, The PIEFII Technology -
Powered by Docking Study with HyperChem THE IMPOSSIBLE BECOMES POSSIBLE.
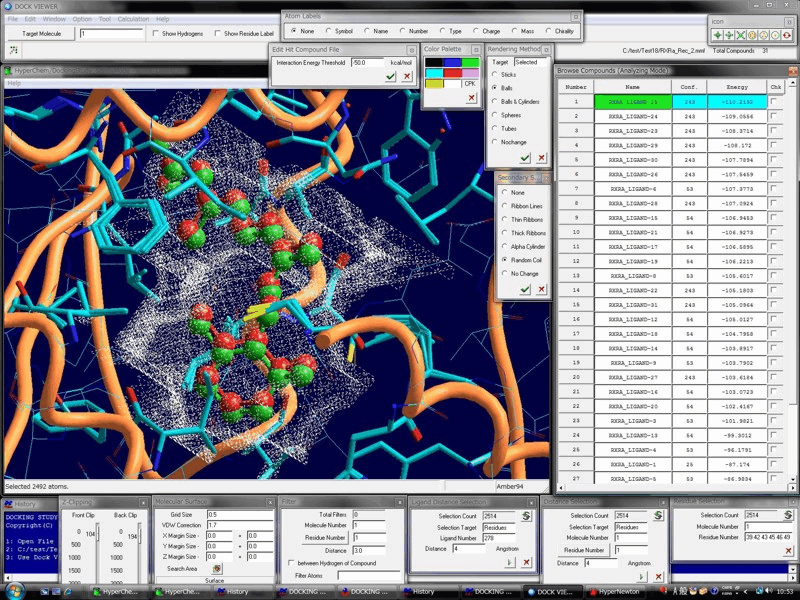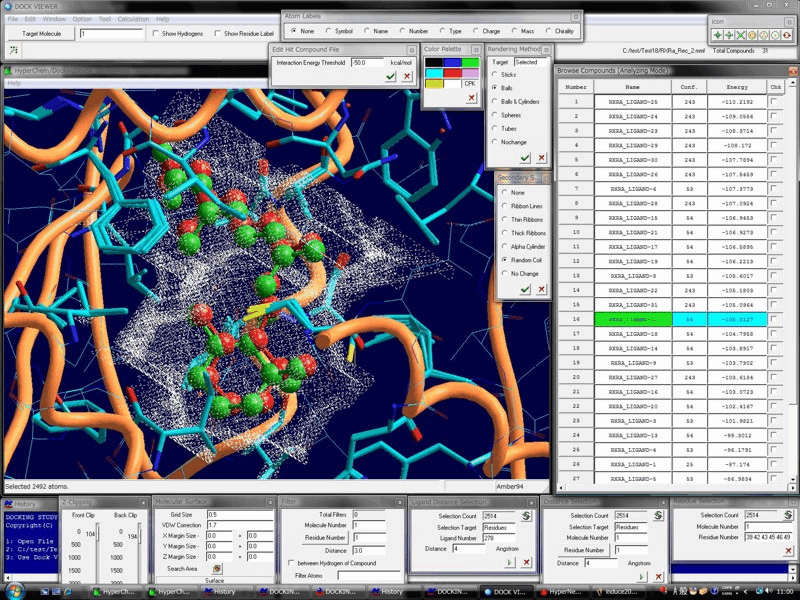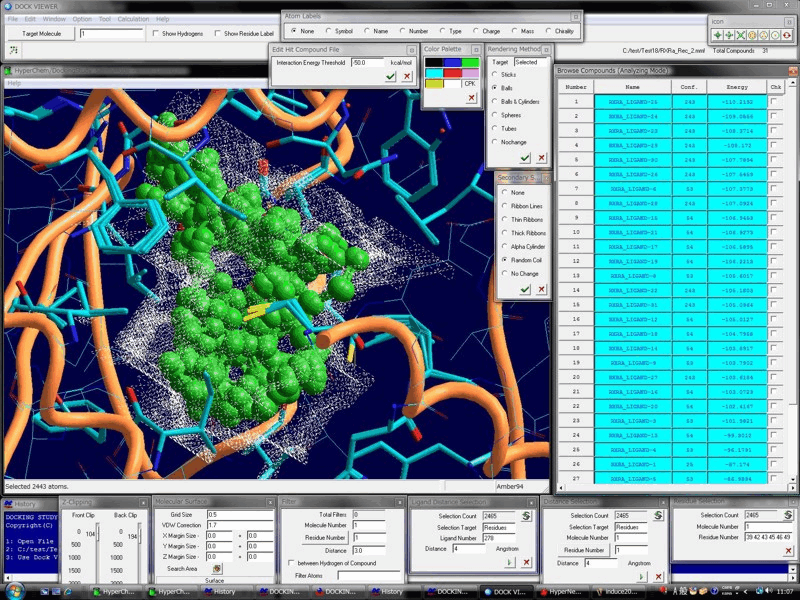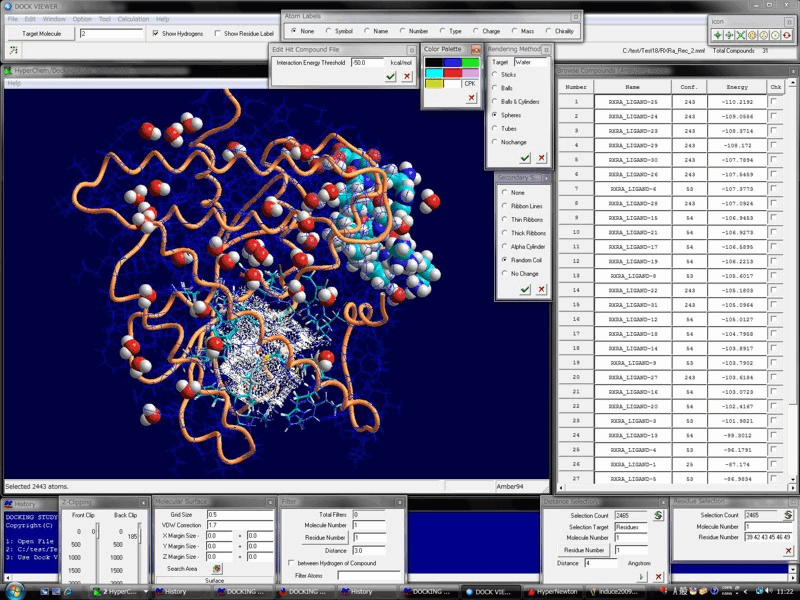
Advanced Technology & The High-Performance, Biomacromolecule- & Ligand-Flexible Docking & Screening Program Package
Docking Study with HyperChem supports the comprehensive and automatic biomacromolecule (protein and nucleic acid molecule systems)- and ligand (small molecule and peptide)-flexible docking simulations, and thus supports the drug design and the in silico screening of a drug candidate.
Homology Modeling Professional for HyperChem, Revision H1
with Gaussian Interface for HyperChem & ONIOM Interface for Receptor
Compatible to NAMD Molecular Dynamics and ABINIT-MP / GAMESS Fragment Molecular Orbital Quantum Mechanics Calculations for Entire Molecular System
Powered by Homology Modeling Professional for HyperChem THE IMPOSSIBLE BECOMES POSSIBLE.
New, High-Performance Software for Biomacromolecular System Modeling, Functional Analysis, and Simulation.
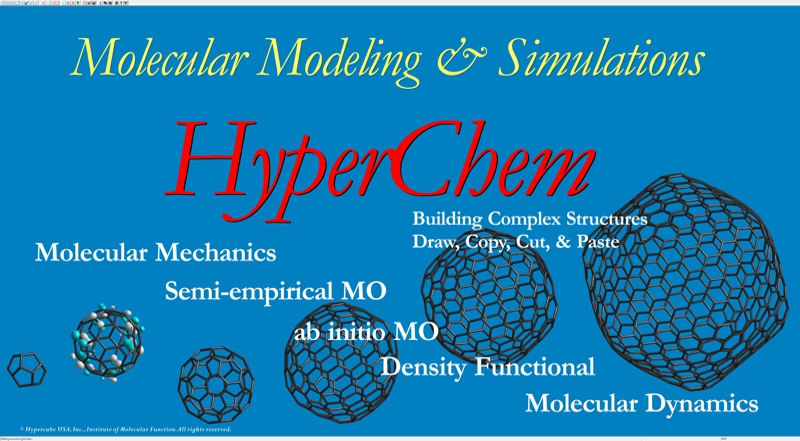
Homology Modeling Professional for HyperChem is the latest molecular modeling package which can carry out the molecular modeling, functional analysis, and simulations of a big molecular system using comprehensively the ab-initio quantum chemistry calculations and the molecular dynamics simulations as well as the molecular mechanics calculations. The program can control seamlessly both HyperChem and Gaussian packages and can be simultaneously fed back their calculation results to the big molecular system in the HyperChem graphical user interface. The whole molecular system can be operated in a quantum chemistry via the full-automatic ONIOM interface to Gaussian. All operations are fully automated in the GUI-based manner, and thus the program supports the logical molecular modeling, functional analysis, and simulations in the life science research such as the structure-based drug design, as well as an unexplored research.
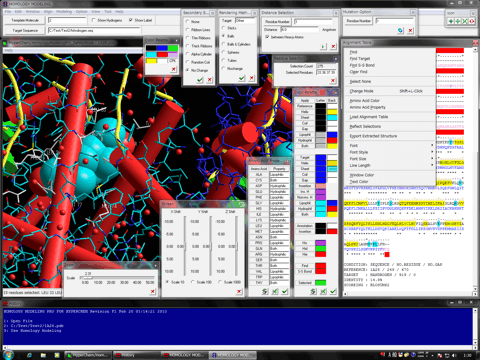
The Next-generation Drug Discovery Technology
PEIFIIPatent Right
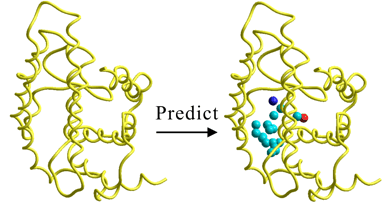
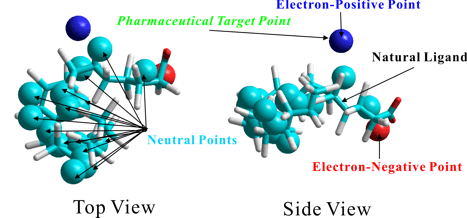
Ab-Initio Biomacromolecule-Ligand Interaction Site (Ligand Binding Site) Prediction Technology
Structure-based Pharmacophore Prediction Technology
Potential Ligand Prediction Technology
Topics
2024/12/15 A research paper between Tokyo University of Agriculture and Technology and a pharmaceutical manufacturer has been accepted by an academic journal International Journal of Molecular Sciences
2024/06/11 Motonori Tsuji ``Evaluation of molecular docking calculations using ab initio dynamics software for drug discovery applications'' was selected for the 2024 Yokohama Industrial Association Research and Development Grant, ``Research and Development Grants for Industry-Academia Collaboration''
2024/04/01 Motonori Tsuji (Principal Investigator), Yokohama National University, Ochanomizu University, and Tokyo Medical and Dental University have been adopted to the 2024th Research Center for Biomedical Engineering (RCBI) project. Structure-based Drug Design for Retinoic Acid Receptor gamma subtype.
2023/12/06 A paper resulting from work with Nagoya University and Shubun University was accepted by an academic journal Microbiology Spectrum
2023/11/01 Institute for Molecular Function begins joint research with Osaka University and Kyoto Prefectural University of Medicine to develop cancer treatment drugs
2023/07/01 20th Anniversary
2023/04/01 Motonori Tsuji (Principal Investigator), Yokohama National University, Ochanomizu University, and Tokyo Medical and Dental University have been adopted to the 2023th Research Center for Biomedical Engineering (RCBI) project. Structure-based Drug Design for Retinoic Acid Receptor gamma subtype.
2022/09/21 Press Release: Potential anti-SARS-CoV-2 drug candidates
2022/09/17 Accepted for publication:Motonori Tsuji. Virtual screening and quantum chemistry analysis for SARS-CoV-2 RNA-dependent RNA polymerase using the ChEMBL database: Reproduction of the remdesivir-RTP and favipiravir-RTP binding modes obtained from cryo-EM experiments with high binding affinity. Int. J. Mol. Sci. 23, 11009, 2022.
2022/05/01 Motonori Tsuji (Principal Investigator), Yokohama National University, Ochanomizu University, and Tokyo Medical and Dental University have been adopted to the 2022th Research Center for Biomedical Engineering (RCBI) project. Structure-based Drug Design for Retinoic Acid Receptor gamma subtype.
2021/10/04 Press Release: Inhibition of main infective pathway of SARS-CoV-2 in oral cavity
2021/09/03 Our research article for COVID-19 inhibition has been accepted for publication in PLOS ONE.
2021/06/23 Motonori Tsuji has contracted the corporate research regarding to retinoids with ChemPhys Co. LTD.
2021/05/14 Motonori Tsuji has been invited to speech at Business Research Institute, 34th CAMM Forum. Development and Application of Structure-based Drug Design Technology - Drug Design for Nuclear Receptor Ligands and COVID-19 medicines.
2021/04/01 Motonori Tsuji (Principal Investigator ), Osaka University, Ochanomizu University, and Tokyo Medical and Dental University have been adopted to the 2021th Research Center for Biomedical Engineering (RCBI) project. Structure-based Drug Design for anti-SARS-CoV-2 drugs.
2020/12/22 Motonori Tsuji (Principal Investigator), Osaka University, Ochanomizu University, and Tokyo Medical and Dental University have been adopted to the 2020th Research Center for Biomedical Engineering (RCBI) project. Structure-based Drug Design for anti-SARS-CoV-2 drugs.
2021/10/17 Motonori Tsuji received the Award of the 31th Japanese Society for Retinoid Research.
2020/09/15 Our resaerch article for acetylcholine esterase inhibitor has been accepted for publication in Chem. Pharm. Bull. journal.
2020/07/02 Our research article (together with Osaka University) for chikungunya virus antibody has been accepted for publication in J. Virol.
2020/05/08 Press Release:Potential anti-SARS-CoV-2 drug candidates
2020/05/01 Research article for potential anti-SARS-CoV-2 drug candidates has been accepted for publication in FEBS Open Bio.Potential anti-SARS-CoV-2 drug candidates identified through virtual screening of the ChEMBL database for compounds that target the main coronavirus protease. FEBS Open Bio, DOI:10.1002/2211-5463.12875.
2019/12/11 Our research article (together with Hirosaki University) for PXR agonist has been accepted for publication in J. Virol.
2018/07/01 15th Anniversary
2017/05/16 Origin of the agonism and antagonism mechanisms in the nuclear receptors via quantum mechanics has been accepted for publication. Motonori Tsuji. J. Comput. Aided Mol. Des. 31, 577-585, 2017, in press, DOI: 10.1007/s10822-017-0025-6.
2017/04/12 Confirmed that the products are compatible to Gaussian16W 64-bit multi-processor version (Revision A.03). I really appreciate great help of Gaussian, Inc.
2017/03/28 All series of HyperChem of Hypercube, Inc. are available from our web site.
2016/12/29 Origin of the receptor subtype selectivity via quantum mechanics has been accepted for publication. Motonori Tsuji, et. al., FEBS Open Bio. 7, 391-396, 2017, DOI: 10.1002/2211-5463.12188.
2016/04/16 Confirmed that the products are compatible to Gaussian09W 64-bit multi-processor version (Revision E.01). I really appreciate great help of Gaussian, Inc.
2015/10/07 Origin of the ligand recognition mechanism in the nuclear receptor superfamily has been published. Motonori Tsuji. J. Mol. Graph. Model. 62, 262-275, 2015.
2015/09/13 A proposed molecular mechanism of ATRA via retinoid X receptor (using core technologies of Docking Study with HyperChem and Homology Modeling Professional for HyperChem) has been accepted for publication. Motonori Tsuji, et. al. J. Comput. Aided Mol. Des. 29, 975-988, 2015.
2015/04/17 Discovery of the underlying rule for the facial stereoselectivity, which is the most important and unsolved issue in organic chemistry, has recently been accepted for publication. Motonori Tsuji. Asian J. Org. Chem. 4, 659-673, 2015.
2014/03/18 A paper regarding the folding mechanism and the agonism and atagonism mechanism of the ligand-binding domain in the nuclear receptor superfamily has recently been published. Motonori Tsuji. J. Struct. Biol. 185, 355-365, 2014.
2013/07/01 10th Anniversary
2012/11/30 Acquired patent rights regarding structural-based pharmacophre prediction technology.
2012/07 Homology Modeling Professional for HyperChem was cited in an article.Plant Cell Physiol. 2012 Jul; 53 (9): 1638-1647.
2010/12 Homology Modeling Professional for HyperChem was cited in an article.Biochemistry 2010 Dec; 49 (50): 10647-10655.
2010/04/30 Homology Modeling Professional for HyperChem was cited in an article.Biochemical and Biophysical Research Communications 2010 Apr 30; 339 (2): 173-177.
2008/02 Docking Study with HyperChem and Homology Modeling Professional for HyperChem were cited in an article. Science 2008 Feb 1; 319 (5863): 624-7.
2008/01/11 Motonori Tsuji was invited as a guest speaker at Business Research Institute.
2007/11/15 HyperChem of Hypercube, Inc. is available as bundle with my products.
2007/06/29 Docking Study with HyperChem and Homology Modeling Professional for HyperChem were introduced in an article. Motonori Tsuji. Molecular Science 2007; 1: NP004.
2007/06/16 I really appreciate great cooperation of Neil S. Ostlund, President and Chief Executive Officer of Hypercube, Inc.
2006/06/05 Docking Study with HyperChem is available.
2005/12/01 Homology Modeling Professional for HyperChem is available.
2005/11/28 Gaussian Interface for HyperChem and ONIOM Interface for Receptor are available.
2005/08/09 My products have been linked to the Gaussian Official site.
2005/07/11 Homology Modeling for HyperChem is available.
2005/05/01 Institute of Molecular Function Web site is started.
2004/03/24 A paper regarding the electronic structure of carboranes and the most stable conformation of cyclopropane ring has been accepted for publication. J. Org. Chem. 69, 4063-4074, 2004.
2003/08/06 A paper regarding carboranyl carbocation has been accepted for publication. J. Org. Chem. 68, 9589-9597, 2003.
2003/07/01 Institute of Molecular Function has been established by Motonori Tsuji.
* HyperChem is a registered trademark of Hypercube, Inc.
*" Gaussian is a registered trademark of Gaussian, Inc.