![]()
HyperChem
Molecular Modeling Software: Cost-Effective De Facto Standard Molecular Modeling System
HyperChem
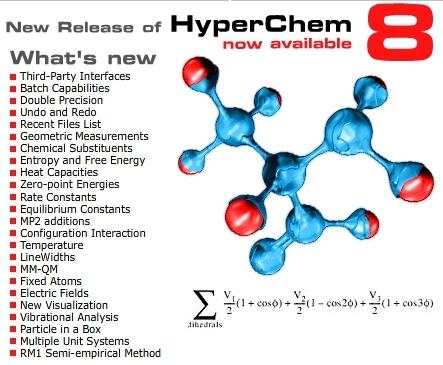
Release 8
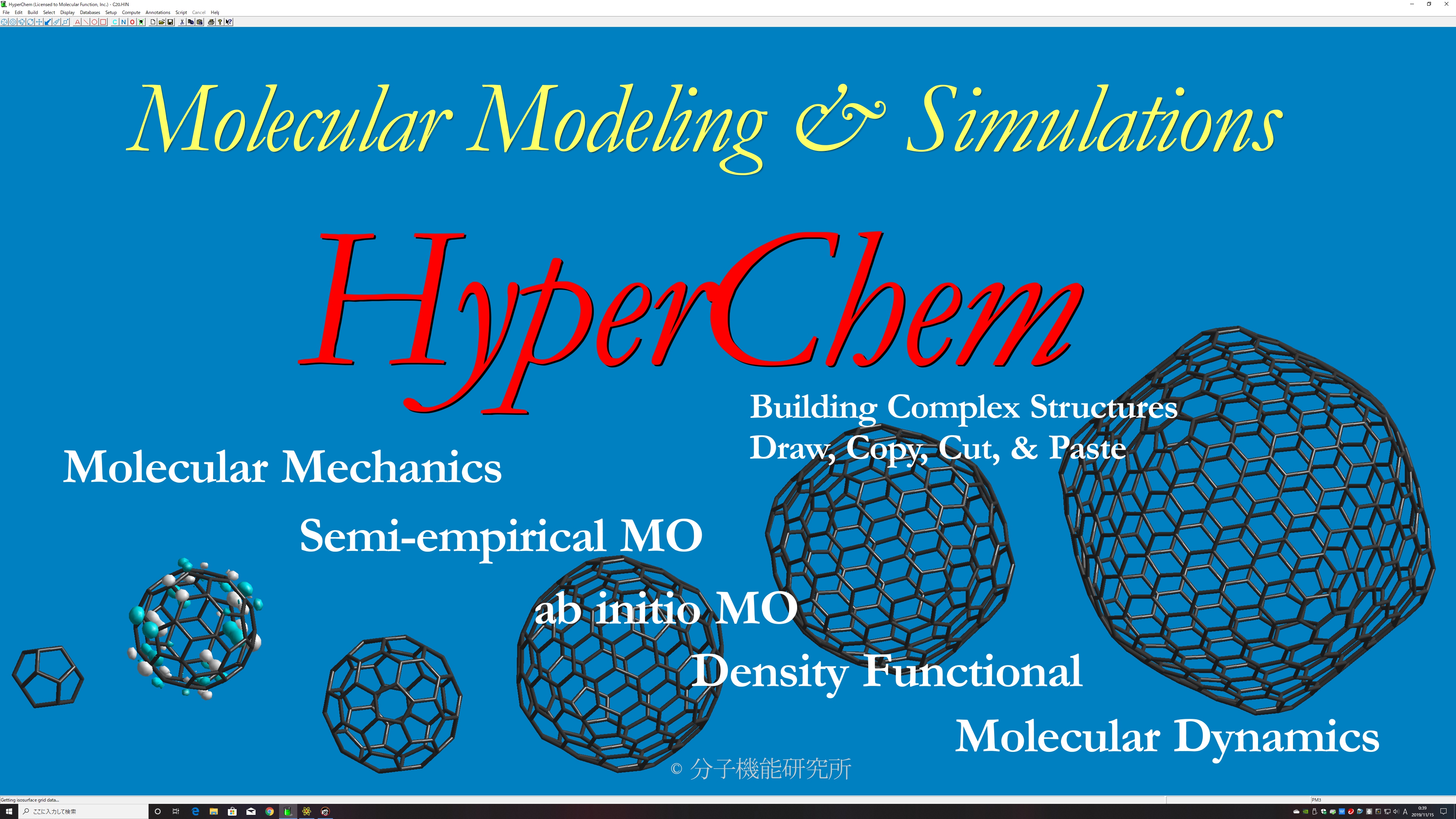
HyperChem is a cost-effective de facto standard molecular modeling software used worldwide by chemists.
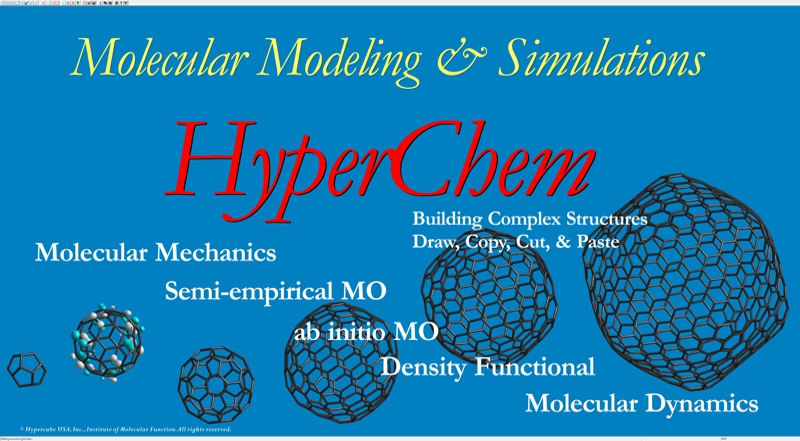
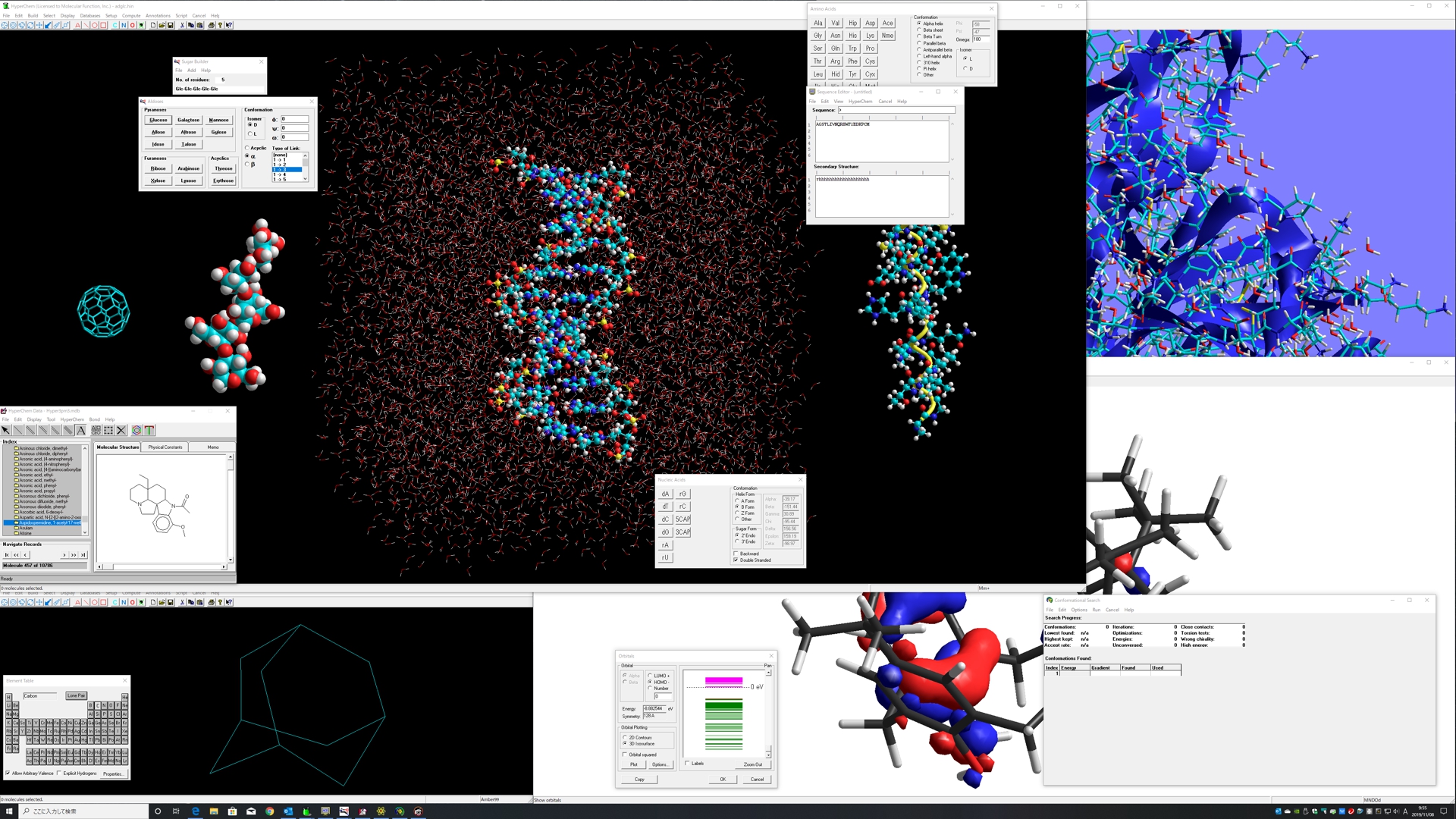
Planar drawing function for any molecules, automatic 3D function, automatic hydrogen addition function, automatic molecular force field assignment function, molecular building function for amino acid, nucleic acid, sugar, and polymer, many types of molecular mechanics, semi-empirical molecular orbital methods, quantum mechanics, density functional methods, molecular dynamics with many types of structural optimization algorithms, effective structure rendering function, scriptable function, and compatibility to many types of file formats.

For more detailed information (HyperChem Special Web Site) (Japaneae)
HyperChem - A Sophisticated Molecular Modeling Environment -
Excellent Molecular Modeling Functions
Unique Molecular Drawing System such as drawing on paper with a pencil
Molecular Cut & Paste Functions
Automatic 3D Function of molecular formula
Automatic Protonation Function
Automatic Force Field Parameter Setting Function
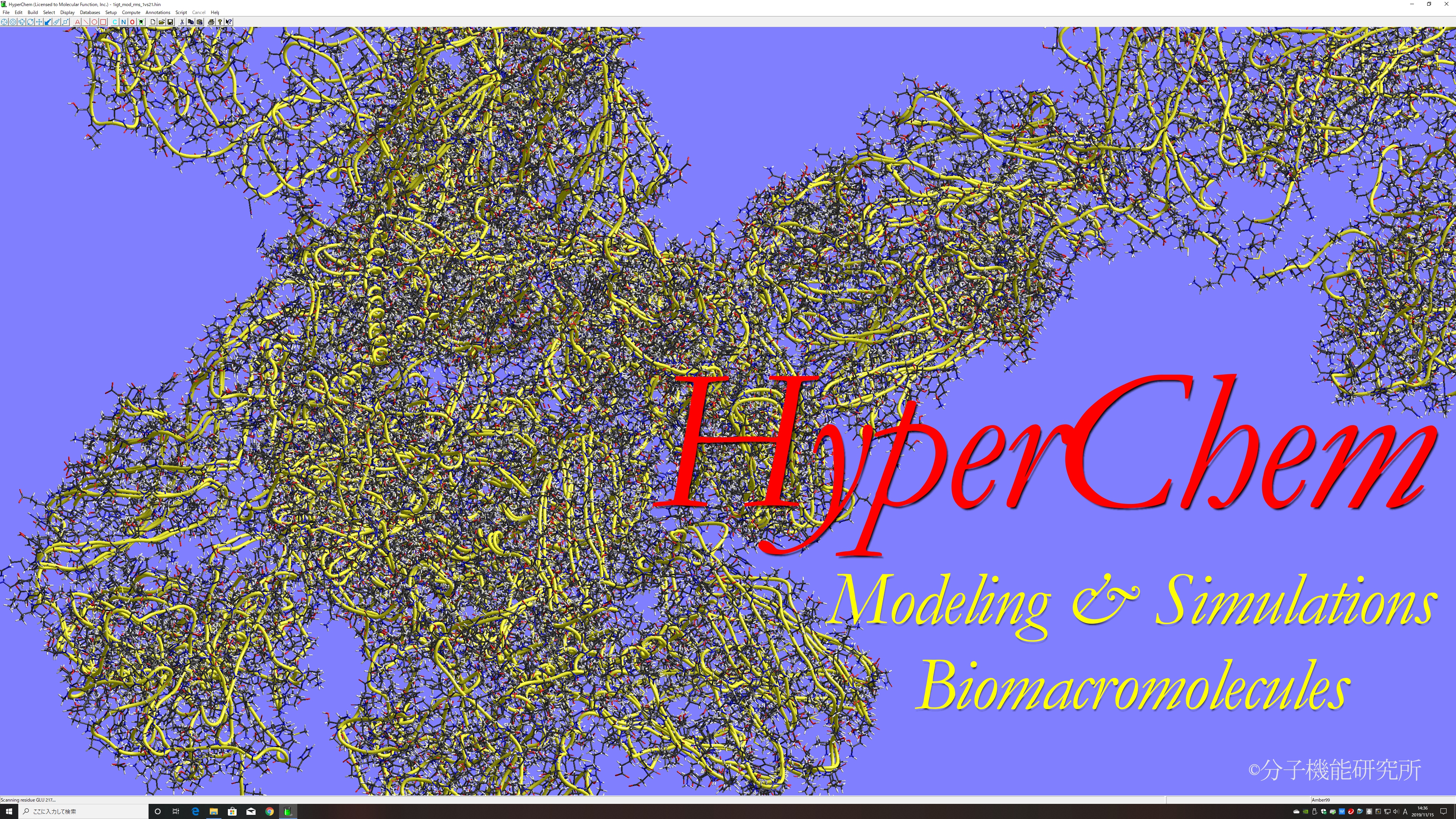
Amino Acid, Nucleic Acid, Sugar, Polymer, and Crystal Edit Functions
RMS Fitting and Molecular Overlay Functions
Conformation Search Function
Comprehensive Computational Chemistry Environment
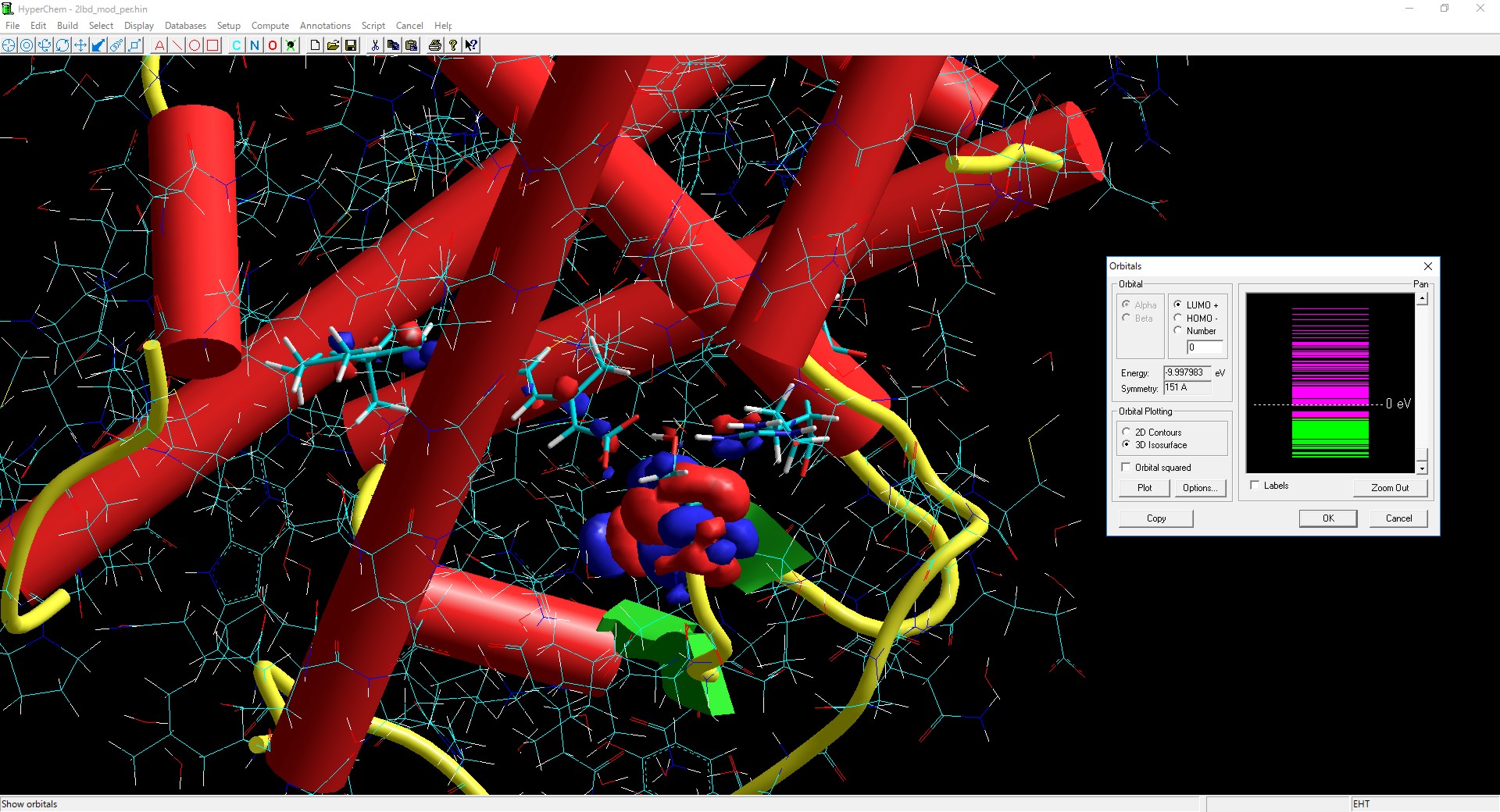
Molecular Mechanics
MM+, Ambers, Amber2, Amber3, Amber94, Amber96, Amber99, Bio+83, Bio+85, Charmm-19, Charmm-22, Charmm-27, Opls, Custom
Semi-empirical Molecular Orbital Theory
Extended Huckel, CNDO, INDO, MINDO3, MNDO, MNDO/d, AM1, PM3, RM1, ZINDO/1, ZINDO/s, TNDO
Ab Initio Molecular Orbital Theory
Hartree-Fock, MP2, CI
Density Functional Theory
Single-Point Calculations
Geometry Optimization Calculations
Molecular Dynamics
Molecular Dynamics, Langevin Dynamics, Monte Carlo
Minimization Algorithm
Steepest Descents, Flecher-Reeves, Polak-Ribiere, Eigenvector Following, Newton-Raphson, Conjugate Directions
Vibration Analysis
Transition State Analysis
NMR Analysis
QSAR
Various Rendering Functions
Selective Display Function
Custom Coloring Function
Labeling Functions
Symbol, Name, Number, Type, Charge, Spin, Mass, Chirality, Basis Set, RMS Gradient, Residue Name, Residue Number, Residue Name+Number, Bond Length, Bond Order, Custom
Molecular Display
Stick, Ball, Ball & Stick, CPK, Tube, Dot
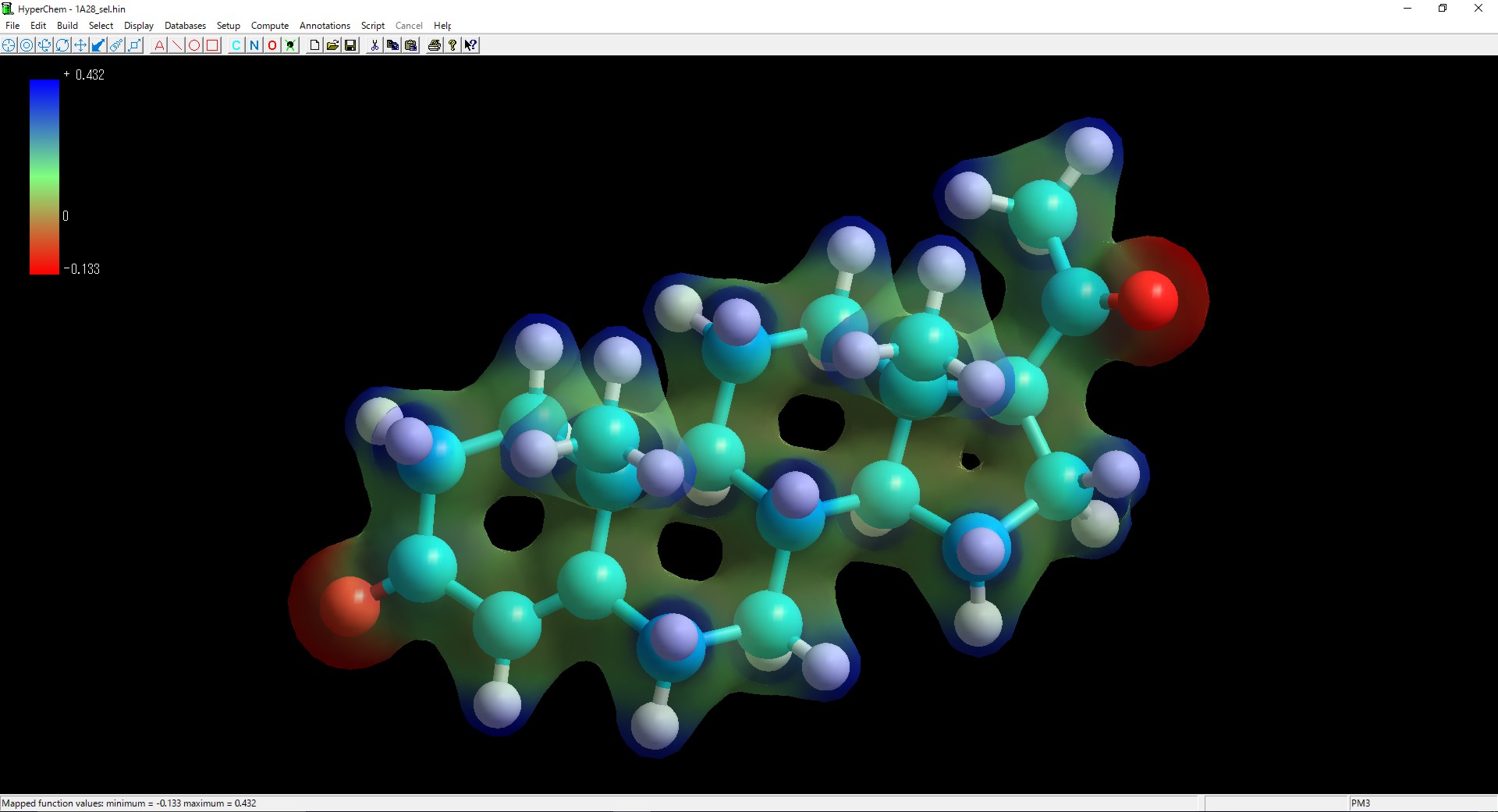
Molecular Orbital, Electrostatic Potential, Electron Density, and Spin Density Maps
2D, 3D, Mesh, Jorgensen-Salem, Line, Flat, Shaded Surface, Gouraud Shaded Surface, Translucent Surface
Hydrogen Bonding
Dipole Moment
OpenGL Secondary Structure and Ribbon Display
Reaction Coordinate, NMR, and Vibration Charts
Development Environment
HCL (HyperChem Command Language) Scriptable
C, C++, Fortran, Tcl/Tk, and DDE Scriptable (for Windows version only)

Compatibility
PDB, ENT, Cartesian, Z-Matrix, ISIS, MDL, TRIPOS, ChemDraw
Recommended Minimum System Requirements
Processor: Intel Core or Xeon (1GHz recommended) (Pentium III, Celeron, or later)
Operating System:
Microsoft: Windows 10 and 11 (32 bit or 64 bit (WOW64) version) (Windows 95 or later)
Mac: MacOS X, OS X, macOS10.x*
Linux:*
Memory (RAM): 2 GB or larger (recommended) (more than 256 MB)
HD: 128 GB or larger (recommended) (300 MB for storage);
Other: CD-ROM drive; Keyboard (touch monitor is also available); Mouse
* Warnings: Mac and Linux versions depend on the version of operating system. Please confirm the compatibility to your environment using Evaluation version.
See also Hypercube, Inc. for details.
Order Information (Customers in Asia)
Please send us HyperChem order form.
HyperChem Full Version for Evaluation
Download site of Evaluation versions
Installation Instruction and Initial Settings
See the following YouTube guide.

* HyperChem is a registered trademark of Hypercube, Inc.