
![]()
News
2020
2020/03/07 In silico screenings targeting SARS-CoV-2 (2019-nCoV) main protease (Japanease).

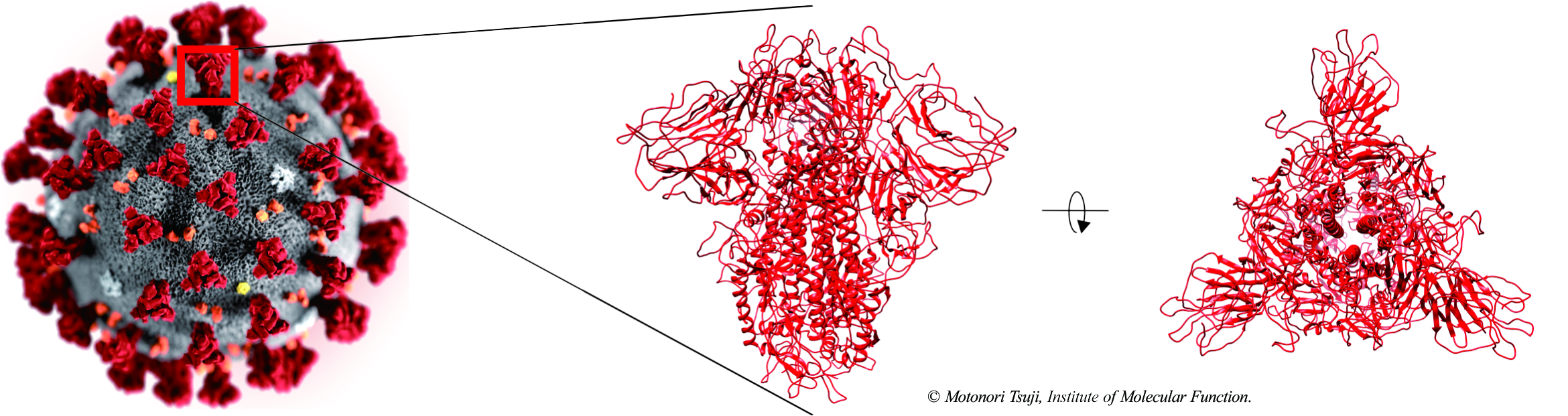
2020/02/25 Antibody design targeting 2019-nCoV spike glycoprotein (Japanease).
2019
2019/12/11
Customer's research article, which Molecular Function has supported the study, has been accepted for publication in J. Pharmacol. Exp. Ther.
2019/11/15
Published the Molecular Function's Loadmap.
Announced the latest version 8.1.3.
2019/11/13
Confirmed that HyperChem Release 8 for Windows, Homology Modeling Professional for HyperChem, and Docking Study with HyperChem are compatible to the latest Windows version (Windows 10 Pro x64, Version 1909).
2019/10/01
Open the Commissioned Calculation Service Web Site (Japanese).
2019/09/01
Research results of MFDD service will be demonstrated at Asian-African Research Forum on Emerging and Reemerging Infections 2019 (2019/09/05-06).
2019/06/01
Confirmed that HyperChem Release 8 for Windows, Homology Modeling Professional for HyperChem, and Docking Study with HyperChem are compatible to the latest Windows version (Windows 10 Pro x64 2019, Version 1903).
2019/02/08
Revised the home pages for mobile devices.
2019/01/26
Open the MFDD Service Web Site (Japanese).
2018
2018/07/01
15th Anniversary
2018/06/07-08
Motonori Tsuji will speak a research entitled "Advancement of the Current Drug Design Technologies via In Silico Drug Design System, Docking Study with HyperChem (DSHC) and Homology Modeling Professional for HyperChem (HMHC)." at the 2018 Spring annual meeting of Society of Computer Chemistry, Japan.

2018/03/27
Motonori Tsuji will speak a research entitled "Antagonist-Perturbation Mechanism for Activation Function-2 Fixed Motifs: Active Conformation and Docking Mode of Retinoid X Receptor Antagonists" at the 138th annual meeting of the pharmaceutical society of Japan.

2018/03/21
Sophisticated Molecular Modeling Software, HyperChem site is started in Japan.
2018/02/23
Announcing Docking Study with HyperChem, Revision H1 and Homology Modeling Professional for HyperChem, Revision H1.
2018/01/09
The Latest In Silico Drug Design Platform: Evolution of Docking Study with HyperChem, Revision H1 and Homology Modeling Professional for HyperChem, Revision H1. Coming Soon. (Strengthened quantum mechanics calculation program, Gaussian, ONIOM Interface, compatible to file format of molecular dynamics calculation program, NAMD (VMD: CHARMM-based PDB), compatible to file format of fragment molecular orbital calculation programs, ABINIT-MP (BioStation Viewer) and GAMESS (Fu /Facio), and supported AutoDock Vina Virtual Screening Interface (preparation of compound database - preparation of all input files - execution - viewing and filtering of hits; supports also simultaneous analysis of both Docking Study and AutoDock Vina results))
2017
2017/11/19
Motonori Tsuji spoke a research entitled "Identifying the Receptor Subtype Selectivity of Retinoids via Docking, QM/MM, and Quantum Mechanics Calculations: ATRA Can Act as an Endogenous RXR Ligand" at the 28th symposium of retinoids of Japan society for retinoid research.
2017/05/16
Origin of the agonism and antagonism mechanisms in the nuclear receptors via quantum mechanics (using core technologies of Docking Study with HyperChem and Homology Modeling Professional for HyperChem) has been accepted for publication. Motonori Tsuji. J. Comput. Aided Mol. Des. 31, 577-585, 2017, DOI: 10.1007/s10822-017-0025-6.
2017/04/16
Confirmed that the products are compatible to Gaussian16W 64-bit multi-processor version (Revision A.03). I really appreciate great help of Gaussian, Inc.
2017/03/28
All series of HyperChem of Hypercube, Inc. are available from our web site.
2017/03/27
Motonori Tsuji spoke a research entitled "Helix H3 Three-Point Initial-Binding Hypothesis: Understanding of Both the Ligand-Trapping Mechanism and the Simultaneous Structural Transition from Apo-Form to Holo-Form of the Ligand-Binding Domain in the Nuclear Receptor Superfamily" at the 137th annual meeting of the pharmaceutical society of Japan.

2017/02/15
Press Release: Origin of the receptor subtype selectivity via quantum mechanics has been published. Motonori Tsuji, et. al. FEBS Open Bio. 7, 391-396, 2017, DOI: 10.1002/2211-5463.12188.
2016
2016/12/29
Origin of the receptor subtype selectivity via quantum mechanics has been accepted for publication.
2016/04/16
Confirmed that ONIOM Interface for Receptor and Gaussian Interface for HyperChem were compatible with the latest Gaussian09W (64bit-multiprocessor version, Revision E.01) under support of Gaussian, Inc. I really appreciate great help of Gaussian, Inc.
2016/03/27
Motonori Tsuji spoke a research entitled "Geometrical Dependence of the Highest Occupied Molecular Orbital in Bicyclic Systems: Origin of the Facial Stereoselectivity of Bicyclic Olefins" at the 136th annual meeting of the pharmaceutical society of Japan.
2016/03/26
Announcing Docking Study with HyperChem 2016 and Homology Modeling Professional for HyperChem 2016, Revision G1.
2016/03/26
The function of ONIOM Interface for Receptor was strengthened (compatible to whole system of protein and nucleic acid molecular system).
2015
2015/12/01
Confirmed that Docking Study with HyperChem and Homology Modeling Professional for HyperChem were compatible with Windows 10 Professional (64 bit Version).
2015/11/04
Press Release: Origin of the ligand recognition mechanism in the nuclear receptor superfamily has been published. Motonori Tsuji. J. Mol. Graph. Model. 62, 262-275, 2015.
2015/10/07
The first success of the structural transition of the holo-form formation simulations and the discovery of the driving forces involved in the ligand entry in the nuclear receptor superfamily using Docking Study with HyperChem and Homology Modeling Professional for HyperChem has been accepted for publication.
2015/09/13
A proposed molecular mechanism of ATRA via retinoid X receptors (using core technologies of Docking Study with HyperChem and Homology Modeling Professional for HyperChem) has been accepted for publication. Motonori Tsuji, et. al. J. Comput. Aided Mol. Des. 29, 975-988, 2015.
2015/04/17
Press Release: Origin of the facial stereoselectivity, which is the most important and unsolved issue in organic chemistry, has recently been accepted for publication. Motonori Tsuji. Asian J. Org. Chem. 4, 659-673, 2015.
2015/03/26
Motonori Tsuji spoke a research entitled "Structure and function of the local motifs in the canonical structure of the ligand-binding domain in the nuclear receptor superfamily" at the 135th annual meeting of the pharmaceutical society of Japan.
2015/01/04
Announcing Docking Study with HyperChem 2015 and Homology Modeling Professional for HyperChem 2015, Revision G1.
2014
2014/10/14
Announcing Docking Study with HyperChem and Homology Modeling Professional for HyperChem, Revision F3.
2014/10/02
Confirmed that Docking Study with HyperChem and Homology Modeling Professional for HyperChem were compatible with Windows 10 Technical Preview (64 bit Version).
2014/03/18
A paper regarding the folding mechanism and the agonism and atagonism mechanism of the ligand-binding domain in the nuclear receptor superfamily has recently been published. Motonori Tsuji J. Struct. Biol. 185, 355-365, 2014.
2013
2013/12/23
Press release (PDF) (Japanese)
2013/12/17
Our paper involved in the breakthrough discoveries for structural biology and bioinformatics has been accepted to publish in Journal of Structural biology.
2013/07/01
10th anniversary
2012
2012/11/30
We have acquired patent rights regarding structural-based pharmacophre prediction technology.
2012/03/04
Announcing Docking Study with HyperChem, Revision F2.
2012/03/03
We were able to confirm that Docking Study with HyperChem and Homology Modeling Professional for HyperChem were compatible with Windows 8 Consumer Preview (64 bit Version).
2012/01/28
Announcing Homology Modeling Professional for HyperChem, Revision F2.
2011
2011/12/01
Important Information about Docking Study with HyperChem and Homology Modeling Professional for HyperChem accompanying minor update of HyperChem 8.0 Professional.
2011/11/13
We were able to confirm that Docking Study with HyperChem and Homology Modeling Professional for HyperChem were compatible with Windows Developer Preview Evaluation Copy Build 8102 (i.e. Windows 8 64 bit Evaluation Version).
2011/09/01
We confirmed a compatibility with Protein Data Bank Format, ver 3.30.
2010
2010/12
Homology Modeling Professional for HyperChem was cited in an article. Biochemistry. 2010 Dec; 49 (50): 10647-10655.
2010/05/16
Announcing changes of the license and price of Docking Study with HyperChem.
2010/04/30
Homology Modeling Professional for HyperChem was cited in an article. Biochemical and Biophysical Research Communications. 2010 Apr 30; 339 (2): 173-177.
2010/03/01
Announcing Homology Modeling Professional for HyperChem and Docking Study with HyperChem, Revision F1.
2009
2009/07/01
Announcing Homology Modeling Professional for HyperChem and Docking Study with HyperChem, Revision E2.
2008
2008/12/01
Altered the licensing system of Docking Study with HyperChem to the perpetual license from the annual license. Simultaneously, changed the restricted number of atoms to 1,000 from 500 in the PIEFII program.
Modified some contents of the upgrade, revision update, maintenance, and support.
2008/11/17
The latest version of Docking Study with HyperChem is available.
2008/11/15
Docking Study with HyperChem, Revision E1 is now available.
2008/10/20
Announcing Docking Study with HyperChem, Revision E1
Docking Study with HyperChem, Revision E1 Pamphlet (PDF: 2 MB; Japanese)
2008/07/29
Free update to Homology Modeling Professional for HyperChem, Revision E1 (version 5.11) is available for customer purchased within one year.
2008/07/27
Homology Modeling Professional for HyperChem Pamphlet (PDF: 4 MB; Japanese)
2008/06/16
The latest version of Docking Study with HyperChem is available.
2008/06/05
Docking Study with HyperChem, Revision D1 is now available .
2008/05/28
SBDD Pamphlet (PDF: 2 MB; Japanese)
2008/05/18
Homology Modeling Professional for HyperChem, Revision E1 is now available.
2007
2007/09/03
Updated the free program package, Molecular Modeling Tools for HyperChem, compatible to the PDB file format version 3.x.
2007/08/30
The latest version of Docking Study with HyperChem is available.
2007/08/28
Ready to ship the latest revisions compatible to the Protein Data Bank format versions 3.0 and 3.1.
2007/08/16
Our products are available from Fiatlux Corporation.
2007/07/01
Updated the free program package, Molecular Modeling Tools for HyperChem, Rev. C1. Please try it on evaluating our products.
2007/06/29
An article entitled Development of the Structure-based Drug Design Systems, HMHC and DSHC has been published in the first issue of Molecular Science electronic journal.
2007/06/16
Homology Modeling for HyperChem Series, Revision C1 compatible with HyperChem8.03 is now available.
Docking Study with HyperChem Series, Revision B1 compatible with HyperChem8.03 is now available.
2007/06/11
Announcing a new version of Homology Modeling for HyperChem.
Announcing a new version of Docking Study with HyperChem.
2007/03/28-30
Docking Study with HyperChem and Homology Modeling Professional for HyperChem in the 127th Annual Meeting of the Pharmaceutical Society of Japan (March 28-30, 2007, Toyama, Japan)
2007/03/16
Updated the free program package, Molecular Modeling Tools for HyperChem. Please try it on evaluating our products.
2007/03/15
New module program, Mol Browser, is added to Docking Study with HyperChem, Revision A3.
2007/03/01
Completed the free upgrade to the corresponding revision B1 for Japanese customers.
The homology modeling tutorials were renewed using the revision B1.
2007/02/21
Homology Modeling Professional for HyperChem, Homology Modeling for HyperChem, ONIOM Inteface for Receptor, and Gaussian Interface for HyperChem Revision B1 are now available for Japanese.
2007/02/14
Workshop in Tokyo (Contents; Japanese)
2007/02/05
Updated the latest pamphlet and release notes for the Homology Modeling for HyperChem package.
2007/01/31
We were introduced to Journal of The International Graph.
2007/01/26
Announcing a new version of Homology Modeling for HyperChem.
2007/01/06
The latest version of Docking Study with HyperChem is available.
2006/12/15
Docking Study with HyperChem supported a novel function which can show some electronic properties such as molecular orbitals, charge densities, and electrostatic potentials for a hit compound complexed to the target protein molecule.
The latest version of Docking Study with HyperChem is available.
2006
2006/12/06-08
Docking Study with HyperChem and Homology Modeling Professional for HyperChem were exhibited in MBSJ 2006 Forum at Nagoya.
2006/11/24
Docking Study with HyperChem was introduced to the Chemical Daily.
2006/11/16
Docking Study with HyperChem, Revision A3 with a multiple-compound screening function is now available.
Press Release (Japanese)
2006/10/27
Announcing Docking Study with HyperChem, Revision A3 with a multiple-compound screening function.
The latest version of Docking Study with HyperChem is available.
2006/10/23
The latest version of Docking Study with HyperChem is available.
2006/10/18
A free package, Molecular Modeling Tools for HyperChem, is available. Please try it on evaluating our products.
2006/10/16
The latest version of Docking Study with HyperChem is available.
2006/10/02
The latest version of Docking Study with HyperChem is available.
2006/09/26
Uploaded a new page, "A New Logical Drug Design Methodology for Organic Chemists". (Japanese language only)
2006/08/18
Our products are available from Scientific Software Service.
The latest version of Docking Study with HyperChem is available.
2006/08/16
The latest version of Docking Study with HyperChem is available.
2006/08/10
Added a series of functions for the molecular surface to Docking Study with HyperChem.
2006/08/05
The latest version of Docking Study with HyperChem is available.
2006/08/03
Docking Study with HyperChem Revision A2 is now available.
2006/07/21
The latest version of Docking Study with HyperChem is available.
2006/07/18
The latest version of Docking Study with HyperChem is available.
2006/07/16
Docking Study with HyperChem and Virtual Screening System now support the restart function.
2006/07/10
Docking Study with HyperChem now carries on a novel function which can update the atomic charges of a trial compound at each conformation using the semi-empirical molecular orbital method.
Virtual Screening System provides a function which can resolve a problem of the rearrangements of the electrons accompanying to the structural changes.
The latest version of Docking Study with HyperChem is available.
2006/07/03
The latest version of Docking Study with HyperChem is available.
2006/06/30
Our products were introduced to the Chemical Daily.
2006/06/29
We were introduced to the Chemical Daily.
2006/06/28
The latest version of Docking Study with HyperChem is available.
2006/06/14
Our products are available from Hulinks Inc.
2006/06/05
Docking Study with HyperChem Revision A1 is now available.
2006/06/01-02
Docking Study with HyperChem and Homology Modeling Professional for HyperChem were exhibited in Society of Computer Chemistry, Japan.
2006/05/30
The PIEFII technology was introduced to the Chemical Daily.
2006/05/08
Our products are available from Lightwave Scientific Ltd.
Press Release (Japanese)
Announcing the Next-generation Drug Discovery Technology
2006/03/08
Developing the Virtual Screening System in the Drug Discovery
Developing the Virtual Screening program based on the Docking Study with HyperChem
Developing the automatic 2D - 3D molecular structure converter, Mol Dimension
2006/03/01
Announcing the Protein-Ligand Docking Study Program, Docking Study with HyperChem b test version.
2006/01/05
The ONIOM Interface for Receptor was introduced to the Chemical Daily.
2005
2005/12/01
Homology Modeling Professional for HyperChem is now shipping.
2005/11/28
ONIOM Interface for Receptor is now shipping.
Gaussian Interface for HyperChem is now shipping.
2005/11/20
Minimum packages including Gaussian Interface for HyperChem and ONIOM Interface for Receptor are available soon.
Homology Modeling Professional for HyperChem is available soon.
2005/11/14
Announcing the full-automatic, multi-layer ONIOM interface, ONIOM Interface for Receptor.
Announcing the molecular modeling and editing program with ONIOM Interface for Receptor, Peripheral Modeling Professional.
2005/11/6
Added a page for the structure-based drug design, the nuclear receptor agonists and antagonists, using Docking Study with HyperChem..
2005/10/31
We were introduced to the Chemical Daily.
2005/10/11
Control Center version 1.03 is available for Homology Modeling for HyperChem.
2005/10/05
Homology Modeling for HyperChem is available from CyberChem, Inc.
2005/08/29
Added a logical molecular modeling tutorial using Peripheral Modeling program and Gaussian Interface for HyperChem.
2005/08/28
Announcing the Latest Protein-Ligand Docking Study Program, Docking Study with HyperChema test version.
2005/08/22
Press Release (Japanese)
Announcing Gaussian Interface for HyperChem.
2005/08/07
Homology Modeling for HyperChem is now shipping.
Gaussian Interface for HyperChem is available for the Peripheral Modeling 1.02 program.
2005/07/26
New module program, Peripheral Modeling 1.x, is added to Homology Modeling for HyperChem, Revision A3.
2005/07/11
Press Release (Japanese)
Announcing Homology Modeling for HyperChem, Revision A3.
Trajectory Analyzer version 1.00 is available for Homology Modeling for HyperChem.
Control Center version 1.01 is available for Homology Modeling for HyperChem.
2005/06/20
Integrated Interface, Control Center, is available for Homology Modeling for HyperChem.
2005/06/13
New Rotamer Database is available for Homology Modeling for HyperChem Revision A2.
2005/06/06
Technical Reports: Side Chain Rotamer Modeling Using Human Androgen Receptor Ligand-Binding Domain.
White Papers: Calculation Times Required for Single-Point Calculations and Geometry Optimizations.
2005/05/31
Test Information: PDB Data set for Homology Modeling for HyperChem, Rev. A2.
Homology Modeling Tutorials Using Homology Modeling for HyperChem, Rev. A2.
2005/05/01 Start Home Page of Institute of Molecular Function .













