
![]()
In Silico Screenings Systems
AutoDock In Silico Screenings Interface
- Docking Study with HyperChem can carry out the virtual screening via AutoDock Vina -
Mol Dimension module program in Docking Study with HyperChem has AutoDock Vina In Silico Screenings Interface, which can automatically construct a compound database in AutoDock PDBQT format from SDF and MOL2 formats and continuously screen the compound database using AutoDock Vina program.**** Thus, combination between Docking Study program and AutoDock Vina program can be used for effectively obtaining a drug candidate.
Preparation of compound database for AutoDock Vina virtual screenings or docking simulations can be performed using Mol Dimension module program.
A configuration file for performing AutoDock Vina can be prepared by PIEFII module program.
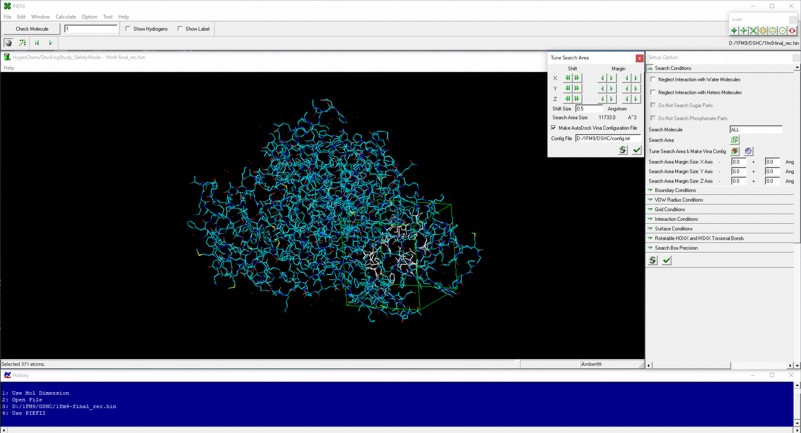
Execution of AutoDock Vina virtual screenings and collection of the resulting output files can be performed by Mol Dimension module program.
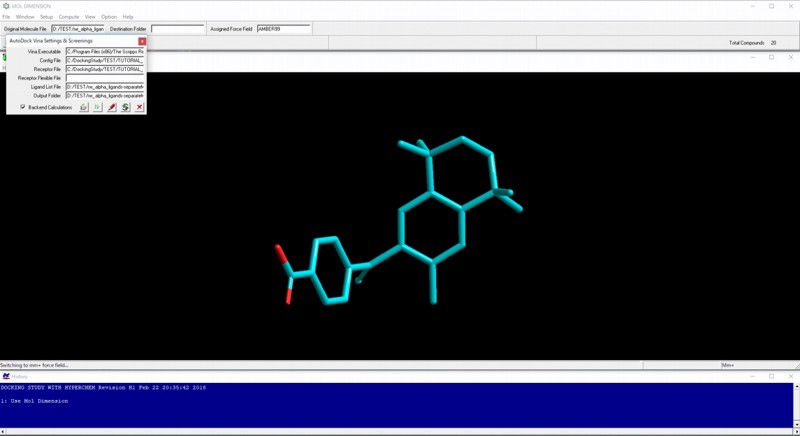
Dock Viewer module program can be used for viewing, analyzing, and filtering AutoDock Vina results.
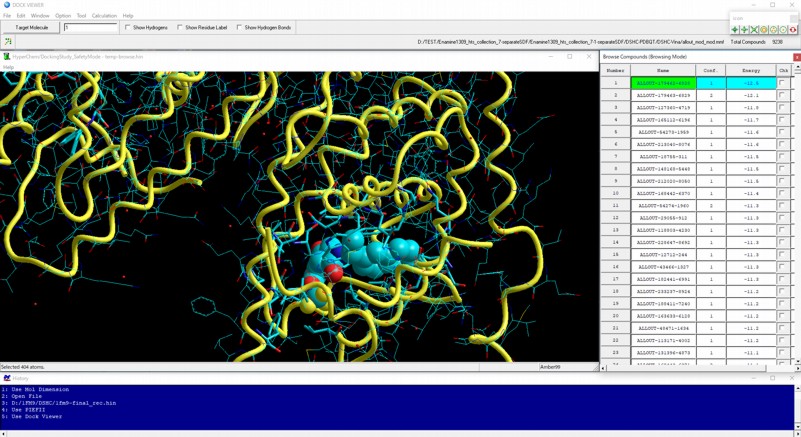
AutoDock Vina in silico (virtual) screenings can be used for million number of compounds. Then, the highly precise docking simulations for the resulting compounds using Docking Study module program are useful for obtaining a drug candidate. In Docking Study with HyperChem, all function related to AutoDock Vina virtual screenings adopts a distributed processing system, so that these operations can be performed by backend calculations using other Windows or Unix machines.
Since Docking Study with HyperChem is also the most cost-effective, it can be expected that this system is useful for learning docking simulations using AutoDock Vina program in pharmaceutical university.
The results of AutoDock Vina screenings can also be viewed using free molecular viewer software such as PyMol.
**** Note that AutoDock Vina program is different docking program from Docking Study with HyperChem. Docking Study with HyperChem only provides the useful GUI environment for performing the AutoDock Vina in silico screenings.













