
Last Modified
1 January 2025
![]()
Homology Modeling Professional for HyperChem
Click on the individual program names to access to their detailed features.
![]()
Protein Superposition
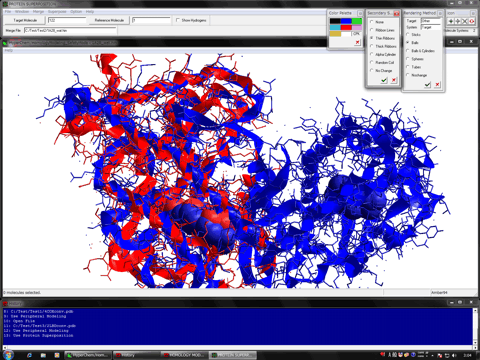
Feature
The "Protein Superposition" program (RMS fit) can automatically superpose merged target molecules (system B) onto the reference molecules (system A) currently loaded into the HyperChem workspace. A molecule system B (and other molecule systems C, D, ...) can be repeatedly superposed onto a desired molecule in the HyperChem workspace (Stacking function) as well as can be unmerged. This program can be used for extraction of important elements from a template molecule system and for constructing a chimera model, as well as used for many structural informatics such as a structural classification of proteins and an analysis of induced fit effect.













