
Last Modified
1 January 2025
![]()
Homology Modeling Professional for HyperChem
Click on the individual program names to access to their detailed features.
![]()
Peripheral Modeling Professional
Feature
In general, a PDB data of a protein crystal structure contains several types of molecules such as substrates, ligands, metals, and other molecules, other than a protein molecule and a water molecule. However, none of chemical information, e.g., hydrogen atom coordinates, bond types, charges, and atom types, etc., for these molecules are given from the PDB data. Therefore, for a model or a crystal structure, which needs to be improved in the precisions, these molecules must be edited for further studies such as the docking study and functional analysis, because these studies cannot ignore the steric and electronic effects of these molecules on the whole structure.
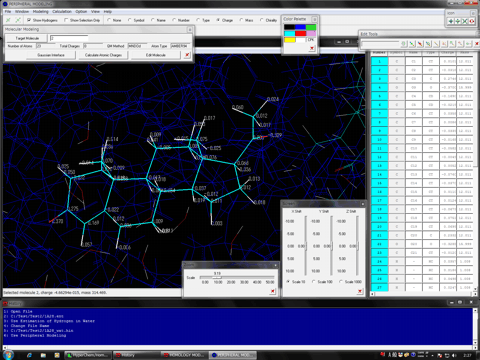
1. The "Peripheral Modeling" program can be used for automatic molecular modeling and editing of small molecules, metals, polymers, metal complexes, and molecules covalently bound to the protein molecule in a protein molecule system. Their chemical properties such as charges and atom types can be automatically assigned under the desired force field conditions using this program. The program can add the hydrogen atoms and bond types to these molecules, as well. User can treat the individual molecules as a single molecule.
2. In general, a small molecule which covalently binds to a protein molecule cannot be treated by the protein modeling. However, the Peripheral Modeling program has solved this problem using an unique QM/MM method, and can simply and precisely make modeling such molecules.
3. The Peripheral Modeling program can automatically add a chemical information which is insufficient in the N- and C-termini and the 5'- and 3'-termini of the original PDB data.
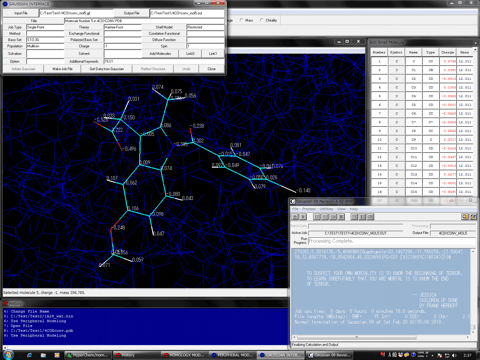
4. The Peripheral Modeling program can automatically and immediately reflect the results calculated by Gaussian program to the corresponding small molecules, metals, polymers, metal complexes, and molecules covalently bound to the protein molecule in the HyperChem workspace using "Gaussian Interface for HyperChem". All hetero molecules in the protein molecule system can be independently and simultaneously subjected to the Gaussian calculations. Thus, this function is useful for a study such as an enzyme-substrate reaction system using the transition state analysis. As the advanced usages, user can reflect atomic charges with consideration of a solvation effect using the reaction field model calculations and can reflect a electronic state of the optimized structure to the corresponding molecules in the HyperChem workspace. The structures optimized by Gaussian can be reflected to the corresponding molecules in the HyperChem workspace using the RMS fitting technique.
5. Moreover, ONIOM Interface for Receptor is available for a molecule system, which consists of biological macromolecules and small molecules such as a receptor and a ligand. ONIOM Interface for Receptor can automatically create a Gaussian job file for the 2-layer (QM:QM and QM:MM) and 3-layer (QM:QM:QM and QM:QM:MM) ONIOM calculations and the result can be reflected to the molecule system in the HyperChem workspace. Thus, as soon as the interface is started, the Gaussian job can be initiated after several seconds without any manual parameter settings, regardless of the preparation of the job file for the 2-layer or 3-layer ONIOM calculation for the big molecular system. The program can reflect some kind of atomic charges and the structures in the optimization processes or the structure optimized.
The fusion of HyperChem and Gaussian programs via these interfaces will be able to provide all of methods for the logical molecular modeling. As a result, the program will be able to support the logical structure-based drug design as well as the logical modeling and the functional analysis of a molecule system.

See the logical ligand design tutorial using the Peripheral Modeling Professional program and Gaussian Interface for HyperChem.













