
![]()
Homology Modeling Professional for HyperChem
Click on the individual program names to access to their detailed features.
![]()
Control Center
Feature
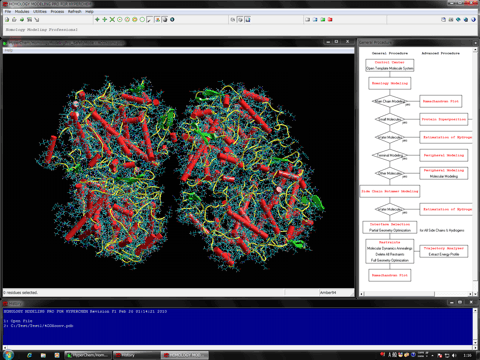
The "Control Center" program can be used for controlling all module programs in Homology Modeling for HyperChem and performing protein homology modeling and functional analysis more efficiently, using HyperChem.
The Control Center serves as a navigator for performing the protein homology modeling with HyperChem, because this program can instruct a homology modeling procedure using the individual module programs and can preserve the operations together with the used file names and program names.
The program also provides the homology search environment. BLAST(New and legacy),ClustalW, and FASTA interfaces are available.
Compatible to the PDB file format version 3.0-3.3.
Molecular system in HyperChem workspace in HIN (HyperChem format) or PDB format can be exported in MDL SDF, Tripos MOL2, and AutoDock PDBQT format using OpenBabel program registered in preference.
The CHARMM-based PDB file extracted from NAMD trajectory can be converted to readable PDB format, so that the equilibrated structure is further modified by molecular modeling functions of Homology Modeling Professional for HyperChem.
Quantum mechanics calculations such as fragment molecular orbital programs, ABINIT-MP (BioStation Viewer) and GAMESS (Fu /Facio) of entire biomacromolecular system can be supported by the conversion function of file format.













