![]()
Homology Modeling for HyperChem
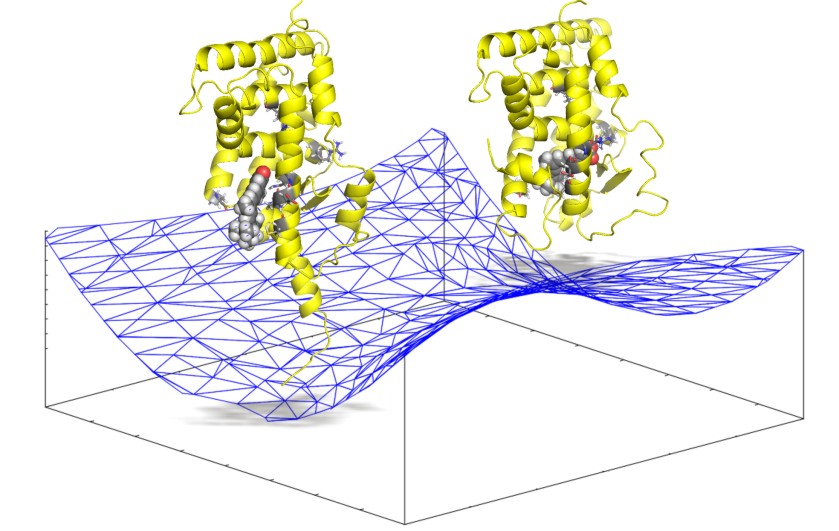
Reaction Coordinate Analysis for Protein-Ligand Complex Formation
Homology Modeling for HyperChem has a powerful potential for performing functional analysis and simulations of protein molecule as well as for performing protein modeling.
For example, the following reaction coordinate in a protein-ligand complex formation can be analyzed in only half a day, using the molecular mechanics calculations.
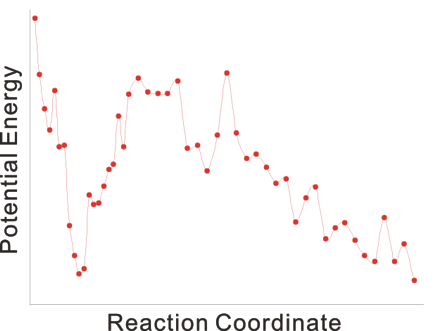
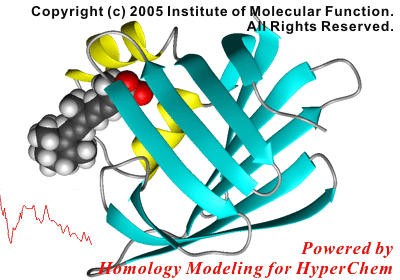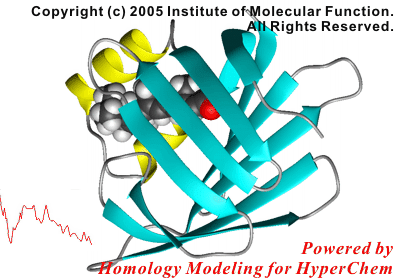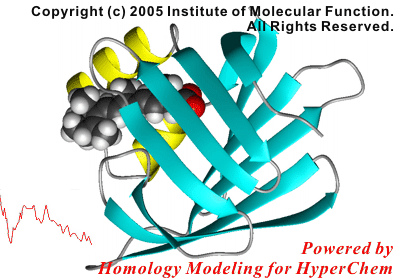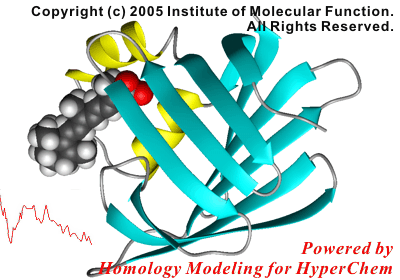
In this study, the initial structure of the protein was modeled using a crystal structure of a homologous, non-liganded protein.
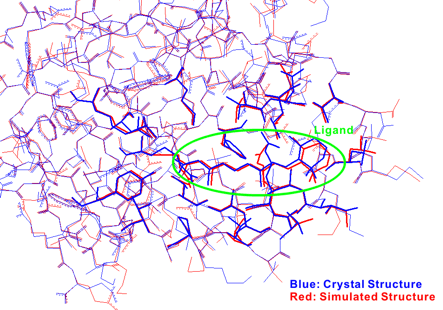
The following figure shows the result of the superposition between the crystal structure and the simulated structure. The results of the present simulations were in excellent agreement with the experimental results, i.e., both the docking mode of the ligand and the side chain conformations contacted to the ligand were reproduced by the present study.
May 2005
for Nuclear Receptor Ligand Recognition Mechanism