Last Modified
1 January 2025
![]()
Bioinformatics
Structural Bioinformatics
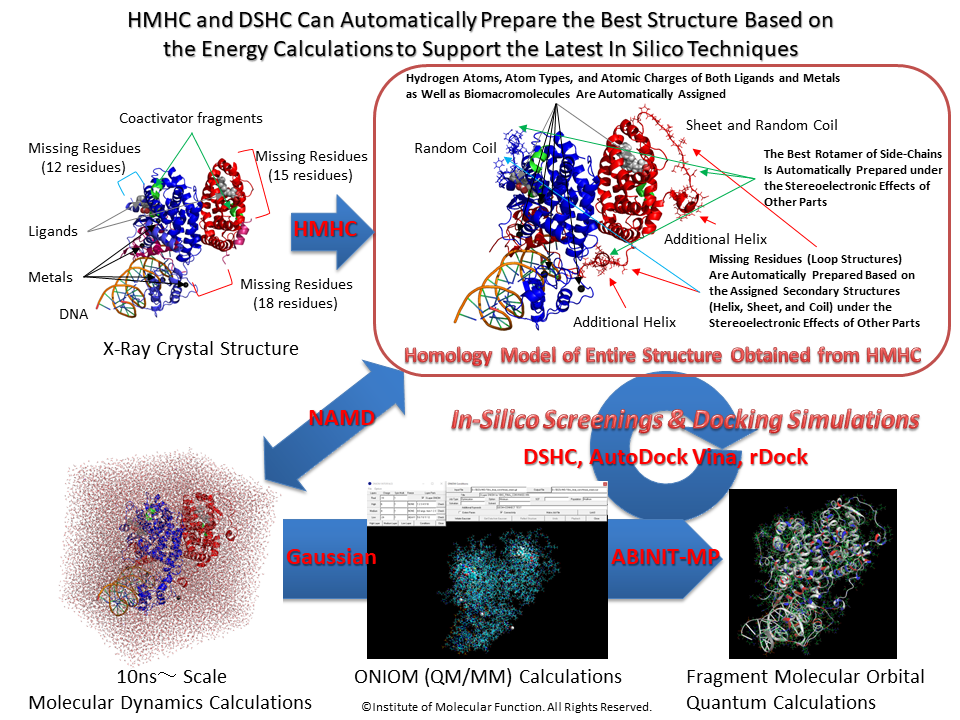
The "ab initio 3D modeling" based on the high precision of our secondary structure prediction algorithm
The predicted structures well reproduced the corresponding crystal structures (refer to the results of the protein superposition). In particular, our algorithm precisely predicted both a loop structure and a random coil structure. Consequently, the secondary structure parts (helices and sheets) are coordinated to a suitable orientation, thus giving a well-reproduced whole structure.
Secondary Structure Prediction
Novel secondary structure prediction algorithm based on the f and y values of main chain atoms
This algorithm shows the high precision in the known algorithms (e.g., Chou-Fasman and neural network algorithms, etc.).
Reported at November 2002.
The Homology Modeling Professional module program in Homology Modeling Professional for HyperChem software carries this secondary structure prediction algorithm and the ab initio 3D modeling algorithm.
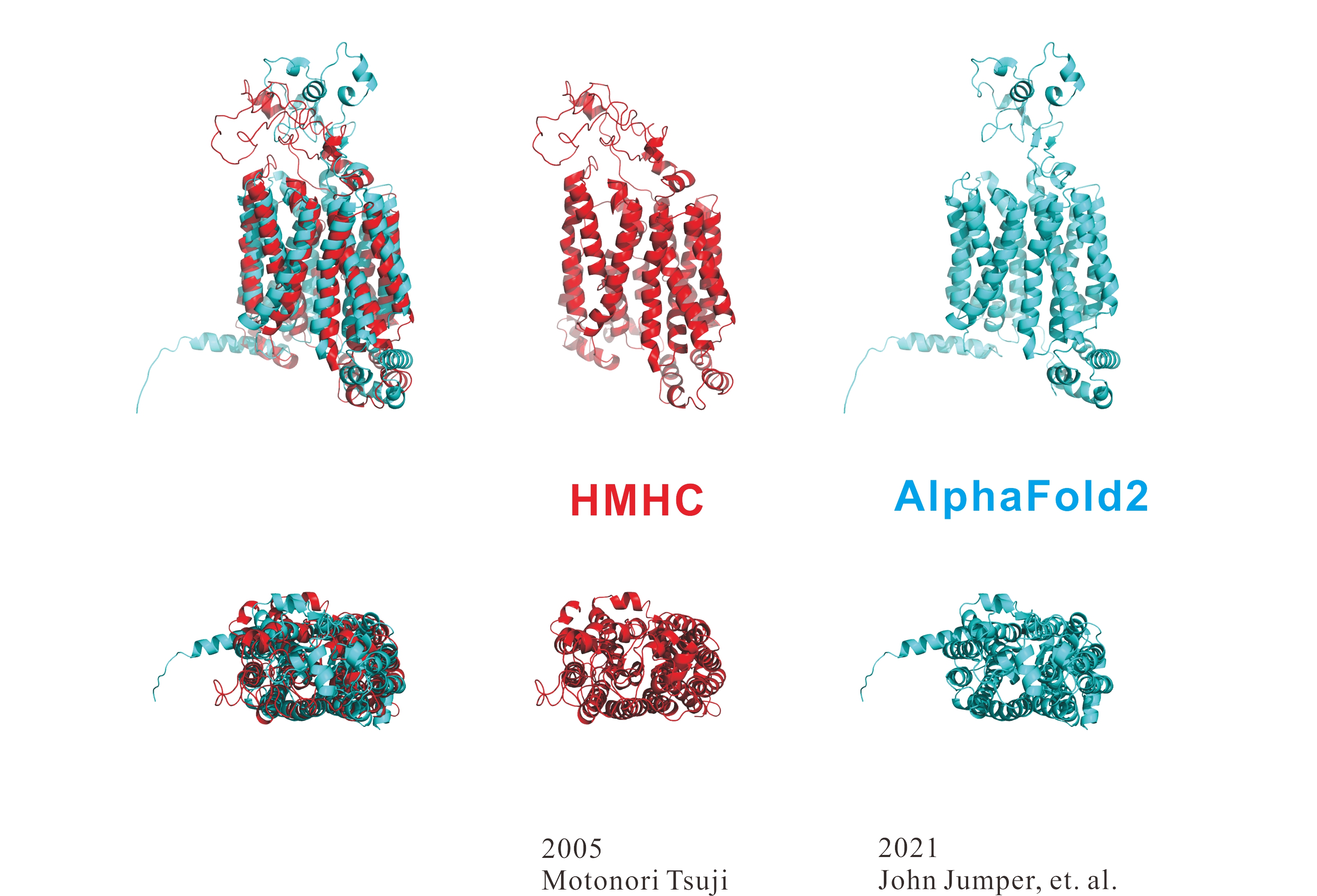
AlphaFold2 vs Homology Modeling Professional for HyperChem (HMHC)
The following figure shows the results of three-dimensional structure prediction for an unknown protein sequence for 20 years including family proteins, using HMHC and AlphaFold2.
HMHC can provide a precise three-dimensional structure since HMHC can model the side-chain rotamer conformations and the interactions between other bioamacromolecules and small molecules as well as crystal waters.