
![]()
Homology Modeling for HyperChem
Homology Modeling Professional for HyperChem, Homology Modeling for HyperChem, ONIOM Interface for Receptor, and Gaussian Interface for HyperChem
What's New in Revisions B1 and C1.
Homology Modeling for HyperChem, Revision C is compatible with HyperChem8.0.
Renewed the interface of all thirteen module programs.
Supported the OpenGL function of HyperChem in all renewed module programs.
Carried the full-automatic rendering and labeling functions.
All advanced functions became more convenient.
Added many useful functions to all renewed module programs.
Carried all theoretical calculations supported by HyperChem.
Removed a functional limitation from the normal version.
In revision B1, normal version is available for all functions and options except for only
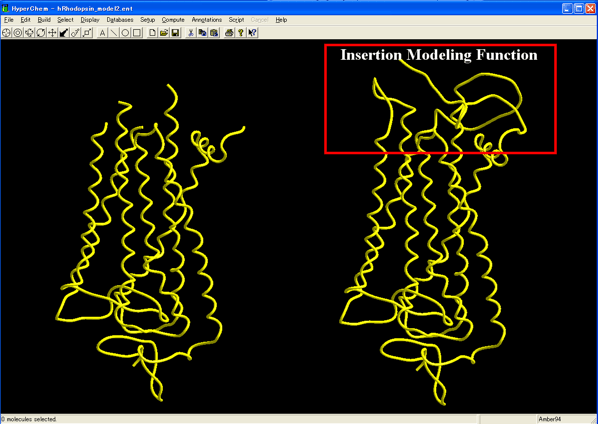
the Insertion Sequence Modeling function in the Homology Modeling Professional module program,
the ONIOM Interface option in the Peripheral Modeling Professional module program, and
the Batch Calculation function for predicting the best rotamer in the Side-Chain Rotamer Modeling Professional module program.
Other Major Additional Functions
Added the novel Insertion Sequence Modeling function to the Homology Modeling Professional module program.
The Side-Chain Rotamer Modeling Professional module program supported a batch calculation for all side chains.
ONIOM Interface supported a protein molecule which covalently binds to a small molecule.
Compatible to the Protein Data Bank file format versions 2.3 and 3.0-3.3.
Revised the user's manual.
Refer to the individual module program pages.
Click on the individual program names to access to their detailed features.
![]()













